Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Granulocytes (neutrophils, eosinophils, and basophils) are short-lived cells that are critical to both antimicrobial and inflammatory responses. The bone marrow (BM) produces granulocytes, particularly neutrophils, at a prodigious rate to supply the baseline need for circulating cells that survive in the peripheral blood for only 3 to 24 hours. It also has the capacity to sharply increase granulocyte production in response to a wide range of stresses. Granulocyte production is controlled by a variety of cytokines that induce the myeloid differentiation program through carefully orchestrated interactions of multiple general and myeloid-specific transcription factors. Understanding this intricate maturation sequence provides important insights into normal neutrophil responses to infectious, inflammatory, and allergic stresses, as well as into the dysregulation of differentiation that leads to myelodysplasia and leukemia.
Granulocytes differentiate from early progenitors in the BM in a process that takes 7 to 10 days. The cells progress through several identifiable maturation stages, during which they acquire the morphologic appearance and granule contents that characterize the mature granulocyte. The earliest identifiable granulocyte precursor is the myeloblast, a minimally granulated cell with scant cytoplasm and a prominent nucleolus ( Fig. 28.1 ). Transition to the promyelocyte stage is associated with the acquisition of abundant primary granules. Primary granules are found in both granulocytes and monocytes and contain proteins necessary for intracellular killing of microbes. The transition to the myelocyte stage is associated with the acquisition of secondary or “specific” granules, which give the characteristic staining pattern that differentiates neutrophils from eosinophils and basophils.
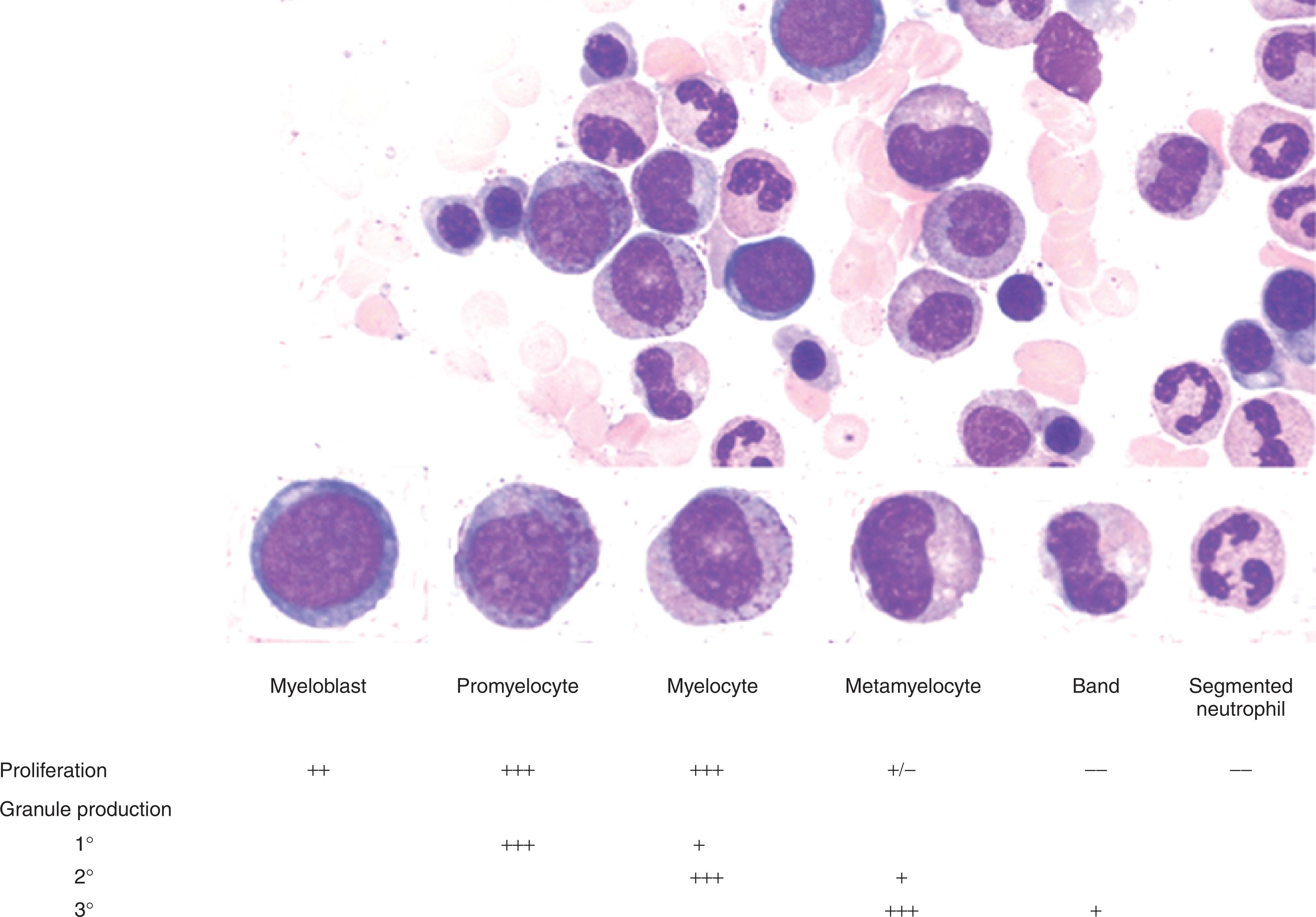
Neutrophil precursors account for approximately half the cells in the BM of normal individuals, with a majority of these at the metamyelocyte stage and beyond. Promyelocytes and myelocytes represent the primary proliferative pool of granulocyte precursors in the BM. Beyond the myelocyte stage, cells mature as nondividing cells. Bands and segmented neutrophils constitute greater than 50% of the total granulocyte mass, primarily as a storage pool of mature cells in the BM. Only 5% of total neutrophils circulate in the periphery, of which 60% are marginated in the spleen and on vessel walls. Mature neutrophils circulate in the peripheral blood for 3 to 24 hours and then migrate to the tissues, where they survive for 2 to 3 days. Therefore the peripheral neutrophil count reflects approximately 2% of the total neutrophil cell mass during approximately 1% of the neutrophil lifespan.
Biochemical events that accompany these physical changes include the sequential acquisition of primary granules and their content proteins (including myeloperoxidase [MPO], lysozyme, neutrophil elastase [NE; encoded by the ELANE gene], defensins, myeloblastin), secondary granules and their content proteins (lactoferrin [LF], neutrophil collagenase [matrix metalloproteinase (MMP) 9], neutrophil gelatinase [matrix metalloproteinase (MMP) 8], neutrophil gelatinase-associated lipocalin [NGAL], transcobalamin 1), and tertiary granules containing neutrophil gelatinase ( Table 28.1 ). The progressive gain of these phenotypic changes and granule acquisition is accompanied by a loss of proliferative potential. This carefully coordinated process is disrupted in acute myeloid leukemia (AML), in which a block in the myeloid maturation pathway usually results in the circulation of immature blasts in the peripheral blood.
| Primary (Azurophilic) | Secondary (Specific) | Tertiary |
|---|---|---|
| Microbial Agents | ||
| Lysozyme | Lysozyme | |
| Myeloperoxidase | ||
| Defensins | ||
| Cationic proteins | ||
| Bactericidal permeability–increasing agent (BPI) | ||
| Proteases | ||
| Elastase | Collagenase | Gelatinase |
| Cathepsin G | ||
| Other proteases | ||
| Acid Hydrolases | ||
| N -acetylglucuronidase | ||
| Cathepsins B and D | ||
| β-Glucuronidase | ||
| β-Glycerophosphatase | ||
| α-Mannosidase | ||
| Other | ||
| Kinin-generating enzyme | ||
| C5a-inactivating factor | Lactoferrin | |
| Vitamin B 12 –binding protein | ||
| Plasminogen activator | ||
| Cytochrome b a | ||
| CD11/1B complex a | ||
| Formyl peptide receptor a | ||
| Histaminase a | ||
| Neutrophil gelatinase-associated lipocalin | ||
a These granule constituents are conventionally assigned to the secondary granule, but their exact compartment remains controversial. Some may be located in the tertiary granule or possibly in one of the other, heterogeneous small-granule populations.
Stem cells have been characterized primarily by their marrow repopulating potential, as outlined in Chapter 9 . Early granulocytic progenitors form hematopoietic colonies in vitro and their more differentiated progeny express specific cell surface proteins that are critically important to myeloid differentiation and function. They mediate both the adhesion of precursors within the BM and the vascular adhesion of mature neutrophils needed for normal neutrophil activation. Other proteins serve as receptors that recognize pathogens or as stimulatory peptides that facilitate activation of phagocytosis and killing of organisms. Appropriate expression of these surface proteins plays an important role in normal neutrophil function, and abnormalities of their expression are implicated in a wide range of diseases affecting the neutrophil compartment. For example, congenital abnormalities in the surface expression of integrin proteins are responsible for failure of neutrophil adhesion in leukocyte adhesion deficiency, whereas acquired abnormalities of expression of the same proteins are thought to underlie the abnormal circulation of immature precursors in myeloproliferative neoplasms. These markers also serve to help distinguish among the stages of myeloid commitment and maturation.
The phenotype of the early hematopoietic stem cells is CD34 + CD38 − CD33 − , with absence of lineage-specific markers. The common myeloid progenitor, colony-forming unit–granulocyte–erythrocyte–macrophage–megakaryocyte (CFU-GEMM), is characterized by the coexpression of CD33. CD33 is expressed at high levels on committed myeloid progenitors and on early precursors of both the granulocytic and monocytic lineages. Expression of CD33 wanes with granulocytic maturation and is absent or nearly absent beyond the myelocyte stage. CD33 is a member of the sialic acid–binding immunoglobulin (Ig)-like lectins (siglecs), which mediate cell-cell interactions and cell signaling pathways involved in immune responses and inflammation. This class of receptors recognizes endogenous sialoglycans as “self-associated molecular patterns” (SAMPs), dampening immune responses via cytosolic Src homology-2 (SH2) domain–containing protein tyrosine phosphatase 1 (SHP1) and SHP2 tyrosine phosphatases and the suppressor of cytokine signaling 3 (SOCS3).
Characteristic granulocyte markers acquired as the early myeloid progenitor cells become committed to the neutrophil lineage include CD45RA, MPO, and CD38, all of which are expressed by myeloblasts. Further differentiation beyond the myelocyte stage is associated with expression of CD16, CD11b/CD18 (Mac-1), and leukocyte alkaline phosphatase (LAP), all of which are expressed at high levels in mature neutrophils.
The acquisition of granules and their content proteins is a critical part of the developmental program of the granulocyte. Arising at specific stages of neutrophil maturation, these intracellular and secretory organelles contain many of the requisite enzymes that mediate the oxidative and nonoxidative killing functions of the neutrophil (see Table 28.1 ).
Primary (azurophilic) granules are acquired at the promyelocyte stage and contain a wide array of proteins, including MPO, defensins, cathepsins, and NE. Secondary granules are secretory granules acquired at the transition to the myelocyte stage. Neutrophil secondary granules contain LF, the vitamin B 12 –binding protein transcobalamin I, the metalloproteinases (neutrophil collagenase and gelatinase), and NGAL. With the exception of gelatinase, which is also expressed by monocytes, expression of the secondary granule proteins is restricted within the hematopoietic lineage to neutrophils. Secondary granules and the synthesis of their contents therefore constitute a definitive marker of commitment to terminal neutrophil maturation. As discussed later, characteristic secondary granules are acquired at the same stage by eosinophils and basophils. Tertiary granules, containing primarily gelatinase, are formed during later stages of neutrophil maturation. Secretory vesicles are formed by endocytosis and contain plasma proteins.
On stimulation, the neutrophil first mobilizes secretory vesicles, which contribute their membrane proteins, including abundant integrin receptors, to the plasma membrane. This increases cellular adhesion by upregulating surface integrin expression in response to selectin stimulation or inflammatory mediators. Primary granules fuse with the phagosome and contribute to bacterial killing. Secondary and tertiary granules have multiple functions. Like primary granules, they fuse intracellularly with the phagosome to help promote bactericidal activity. They also contribute membrane proteins to the plasma membrane and are the source of the surface integrin receptor Mac-1 (CD11b/CD18) that is prominently upregulated during neutrophil activation. Secondary and tertiary granules also release the matrix-modifying metalloproteinase collagenase (MMP8) and gelatinase (MMP9) into the extracellular milieu, enhancing neutrophil penetration into sites of inflammation. LF and transcobalamin I are thought to contribute to the antimicrobial response by sequestering iron and cobalamin, respectively, away from infecting organisms. LF has also been implicated in modulating neutrophil extracellular traps (NETs) by inhibiting their release.
The fusion of azurophilic and peroxidase-negative granules allows for cross-exposure of their contents within the phagosome. Under resting conditions, their constituent proteins are carefully sequestered in separate organelles, preventing premature activation and damage to the resting neutrophil. When granule fusion occurs, their protein contents cooperate to generate the antimicrobial response. Hydrogen peroxide, a by-product of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase in secondary granules, combines with MPO from the primary granules to produce hypochlorous acid, a highly toxic microbicidal agent. The secondary granule proteins neutrophil gelatinase and neutrophil collagenase are produced as zymogens and are converted to their active forms by the action of NE released from the primary granules.
Current evidence largely supports the hypothesis that the content of the neutrophil granules is dictated primarily by the timing of synthesis of their respective content proteins. Multiple studies have demonstrated that each distinct granule population is generated not by differences in protein sorting but rather by a highly regulated transcriptional process that results in sequential gene expression. Expression of MPO occurs between the promyelocyte and the myelocyte stages of neutrophil development, and as a result it is packaged into the primary granules. Secondary granule proteins such as LF are expressed between the myelocyte and the metamyelocyte stages and hence are packaged into the secondary granules. Overexpression of the secondary granule protein NGAL in HL60 cells, a leukemic cell line that is arrested at the myeloblast stage of differentiation, results in its incorporation into primary granules, supporting the notion that gene expression and protein sorting into granules are coordinated events. However, this model may be an oversimplification because there is some overlap of expression between certain primary and secondary granule protein genes, and primary granule protein gene expression is much less synchronous than that of secondary granule proteins. The defensins are expressed later than other primary granule proteins, and defensin transcription appears to be regulated by the same transcriptional regulatory pathway as for the secondary granule proteins gelatinase and LF. The defensins are a family of small, variably cationic proteins that are highly abundant in the granules of mammalian phagocytes. Three defensins, HNP-1, -2, and -3, comprise 30% to 50% of total protein in the azurophilic granules of human neutrophils. Some defensins are broadly antimicrobial, antiviral, and cytotoxic, whereas others are chemotactic or opsonic or may modulate hormonal responses. The defensins are the only primary granule proteins that are absent in patients with neutrophil specific granule deficiency (SGD), which would predict targeting to the secondary granule. How defensins are packaged exclusively in the primary granule remains unclear.
Granulocytes arise from pluripotent hematopoietic stem cells by a process of commitment, proliferation, and differentiation. Stem cells are long-lived cells capable of both self-renewal and differentiation to lineage-specific–committed progenitors (see Chapter 9 ). The process governing the cell fate decision that takes a stem cell down the path to lineage commitment and the subsequent factors that regulate lineage-specific differentiation have been the subjects of intense study for many years. Three models have been proposed for the mechanism underlying lineage commitment and differentiation of the pluripotent stem cell. The first, or inductive , model suggests that lineage commitment and differentiation are the results of external stimuli (e.g., growth factors, stroma). The second model, the stochastic model, emphasizes intrinsic cellular factors as being critical to hematopoiesis, and the third model combines the attributes of the first two. It appears that the transition from a stem cell to a committed progenitor is largely stochastic, although subsequent maturation from progenitor to precursor to mature neutrophil requires cytokine activity. Controversy remains as to whether cytokines and the BM microenvironment play instructive or permissive roles in influencing stem cell commitment and in inducing the proliferation and maturation of committed progenitors.
Recent observations in mouse models of hematopoietic stem cell transplantation suggest that neutrophils themselves play active roles in modulating the BM microenvironment by driving sinusoidal regeneration following transplantation. Aging neutrophils reenter the BM as well as extramedullary tissues from the peripheral circulation, and their elimination by resident macrophages activates feedback loops that regulate granulopoiesis through cytokine signaling (reviewed by Cossio et al. ). The interplay between neutrophil life cycle, BM niche, and cross-talk with extramedullary organ systems currently remains an active area of investigation.
Early progenitor cells express receptors for multiple cytokines, but their expression becomes more restricted as cells become committed to a specific lineage. As a consequence of the broad range of cytokine receptor expression, early progenitors respond to combined growth factors, many of which show synergistic activity. The early-acting cytokines include the interleukins IL-1 and IL-6, stem cell factor (SCF), FLT3 ligand, and several others, including granulocyte colony-stimulating factor (G-CSF) and thrombopoietin (TPO). IL-3 is important in directing the pluripotent stem cell toward the myelomonocytic lineage, giving rise to the mixed myeloid progenitor (CFU-GEMM). Subsequent stages leading to commitment- and lineage-restricted differentiation are governed by late-acting cytokines ( Fig. 28.2 ).
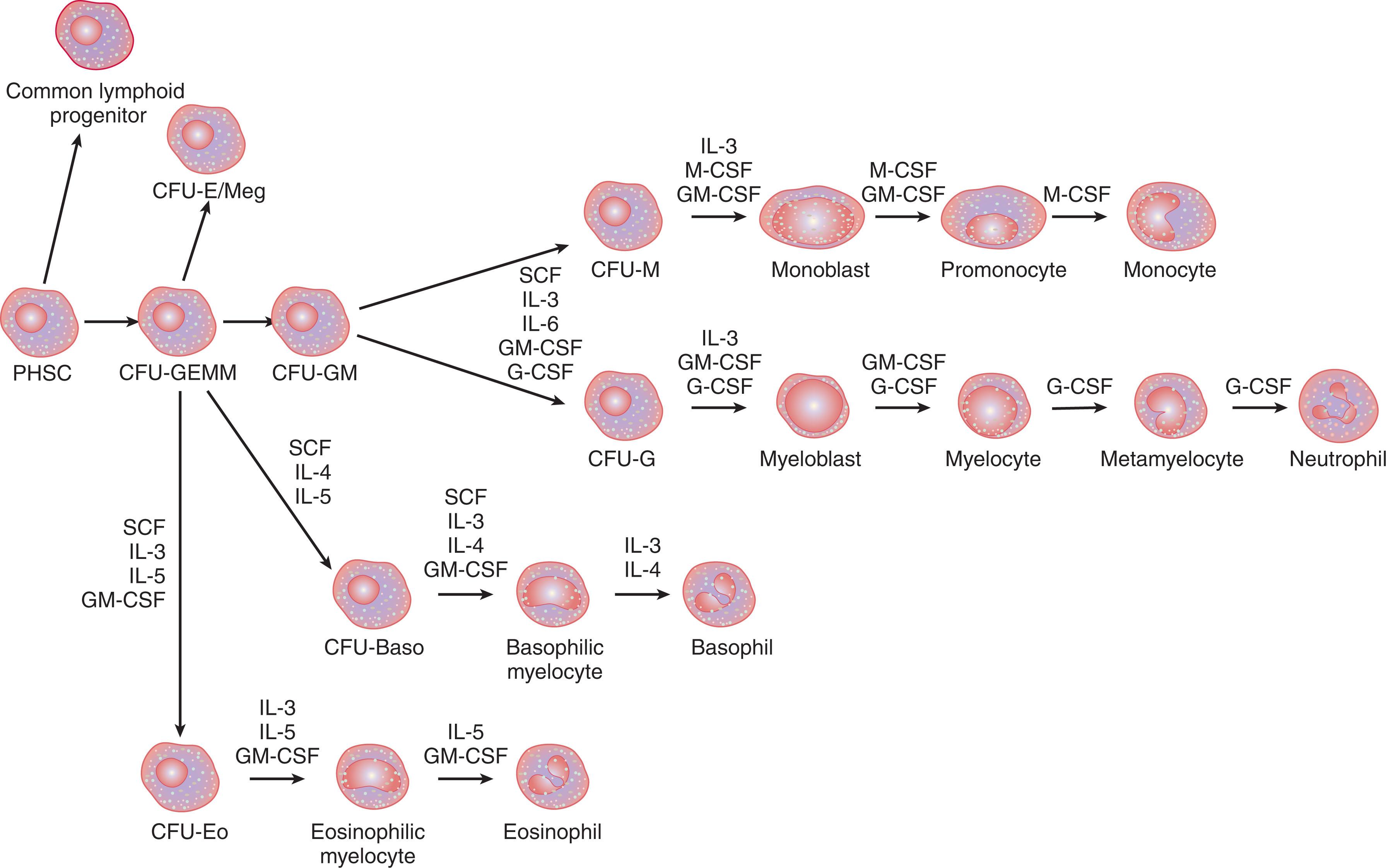
The major cytokines that mediate neutrophil maturation are G-CSF and granulocyte–macrophage colony-stimulating factor (GM-CSF). G-CSF not only supports the survival and proliferation of developing myeloid cells at all stages of differentiation but also increases the functional activity of mature neutrophils. G-CSF binding to its receptor, G-CSFR, leads to homodimerization and signal activation of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) and the Ras/mitogen-activated protein kinase (MAPK) pathways. The role of G-CSF as both an early- and late-acting cytokine is underscored by the successful use of G-CSF to mobilize early progenitors into the peripheral blood for stem cell collection and to speed neutrophil recovery following chemotherapy. Although the major role of G-CSF is thought to be induction of neutrophil proliferation and differentiation, the G-CSFR is also expressed on a wide range of cell types. In addition to myeloid progenitors and precursors at all stages of neutrophil differentiation, the G-CSFR is expressed on platelets, monocytes, and lymphocytes, as well as some nonhematopoietic tissues, including endothelium and placenta.
The importance of G-CSF in myeloid proliferation and differentiation has been studied in G-CSF–null and G-CSFR–null mice. Mice lacking G-CSF or G-CSFR have markedly decreased myeloid progenitors and impaired neutrophil production, with low circulating neutrophil counts. In addition, G-CSF–null mice have impaired neutrophil mobilization, and mature neutrophils from G-CSFR–null mice have increased susceptibility to apoptosis, supporting the role of the G-CSF pathway in sustaining the mobilization, survival, and function of mature neutrophils. Despite these abnormalities, G-CSF/G-CSFR–knockout mice are still capable of some neutrophil production, suggesting that there are alternative cytokine pathways that allow for granulocyte development.
GM-CSF also induces proliferation and differentiation of myeloid precursors. The GM-CSF receptor (GM-CSFR) is a heterodimeric protein composed of an α- and a β-subunit. The α-subunit binds GM-CSF. The β-subunit is shared by GM-CSFR and the receptors for IL-3, IL-5, and IL-6. The β-subunit does not bind ligand but is necessary for high-affinity ligand binding to the αβ-heterodimer of each receptor. Signaling through GM-CSFR activates both the JAK/STAT pathway and the Ras/MAPK pathway. Of note, GM-CSF–null mice have no defect in hematopoiesis, whereas mice with null mutations in both G-CSF and GM-CSF have more profound neutropenia in the perinatal period but the same levels of neutrophils in adulthood as those of mice lacking G-CSF alone.
Lineage-specific maturation of committed hematopoietic progenitors is driven by transcription factors controlling cellular proliferation, differentiation, and survival of the totipotent stem cell, and these roles have been well established with the aid of mammalian animal models. Studies of the regulation of individual genes that show tissue- and stage-specific myeloid expression have implicated a small number of transcription factors that are responsible for directing both phenotypic myeloid maturation and the expression of functionally important myeloid genes. Furthermore, pathognomonic chromosomal translocations resulting in the dysregulation of transcription factor expression and activity, leading to defects in myeloid-specific gene expression, have been well described in AML.
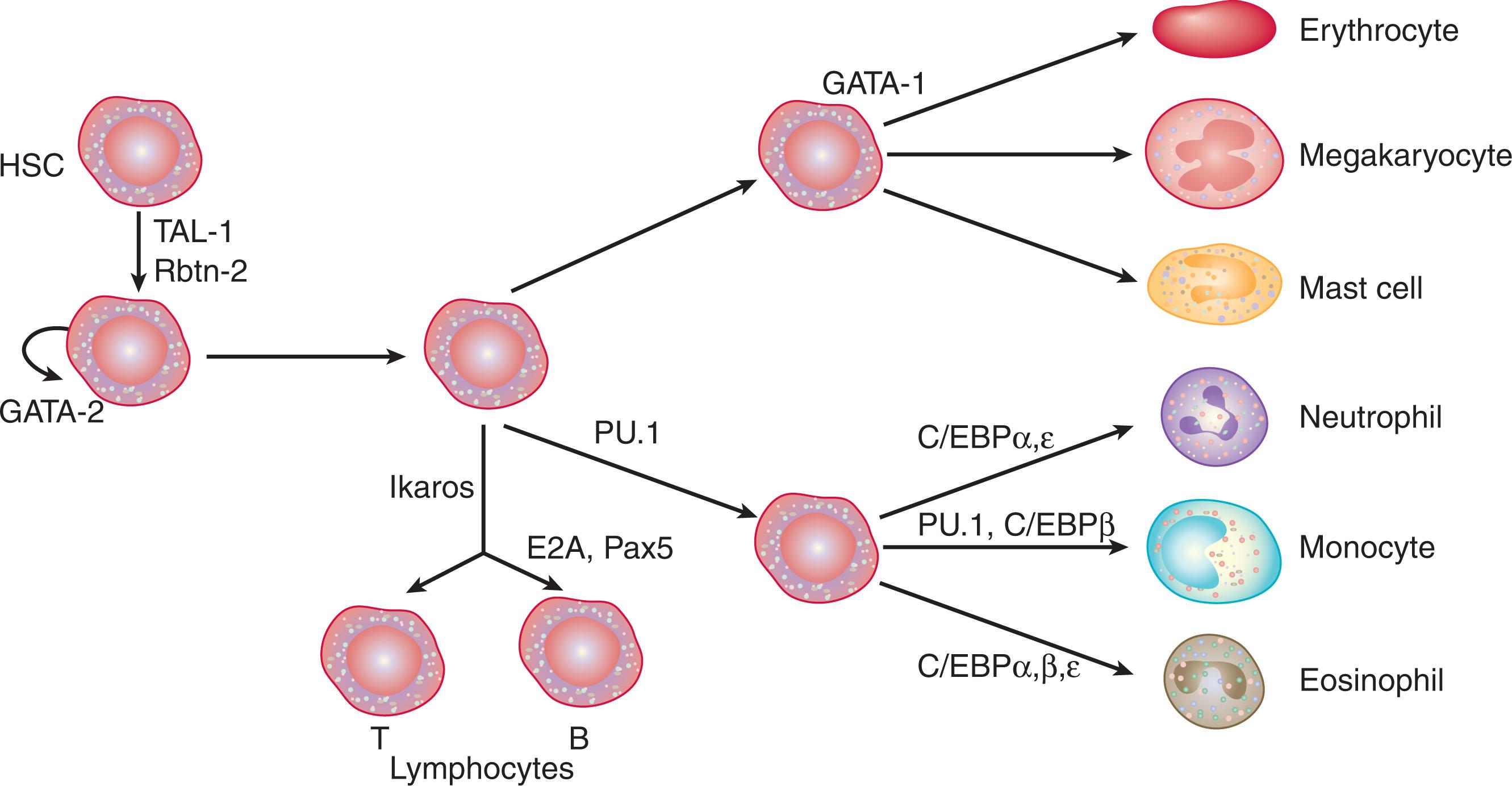
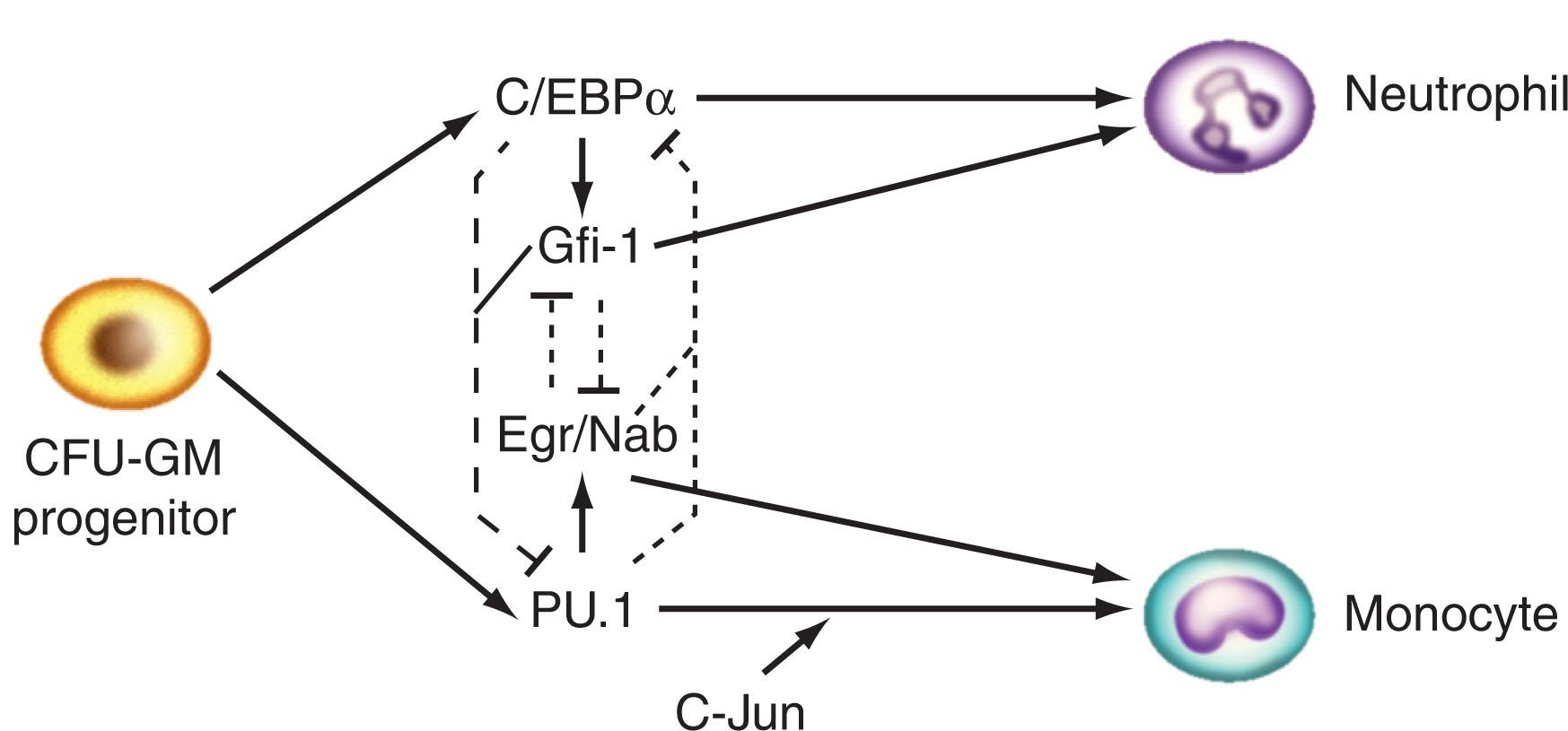
On the basis of experimental observations using genetic knockout techniques, overexpression studies, and multicolor flow cytometry, critical transcription factors have been identified and are classified into two major categories. The first category includes stem cell leukemia transcription factor (SCL), GATA-2, and AML transcription factor-1 (AML-1; now known as Runx1), all of which influence differentiation of all hematopoietic lineages. The second category comprises the master regulators of lineage development, including GATA-1, PU.1, and CCAAT enhancer-binding protein-α (C/EBPα). These factors not only promote lineage-specific gene expression but also suppress alternative lineage pathways. Fig. 28.3 summarizes the postulated role of several key transcription factors during hematopoietic development. Myeloid progenitors exhibit multilineage patterns of transcription factor gene expression. Laslo et al. elegantly demonstrated that cell fate determination is dependent upon subtle changes in expression levels of transcription factors that regulate differential lineage maturation. For example, levels of PU.1 expression are increased by Egr-1/Nab-2 in developing macrophages; at the same time, Egr-1 inhibits the expression of the neutrophil-specific transcription factor growth factor independence-1 (Gfi-1), thereby simultaneously repressing the neutrophil development program ( Fig. 28.4 ).
Runx1, previously known as AML-1, belongs to a family of highly conserved transcription factors that harbor a 128-amino-acid motif referred to as the Runt domain. The Runt domain functions in DNA binding, protein-protein interaction, adenosine triphosphate (ATP) binding, and nuclear localization. This family of transcription factors, known as the core binding factor (CBF) family, has been implicated in specification of cell fate and has roles in myeloid differentiation and lineage-specific granulocytic function.
Runx1 is the DNA-binding α-subunit of the CBF complex. Together with CBFβ, a widely expressed protein that enhances the DNA-binding affinity of the α-subunit, Runx1 binds the consensus DNA motif 5′ Pu ACCPuCA 3′ as a dimer. Disruption of the Runx1 gene in mice results in embryonic lethality due to failure of definitive hematopoiesis in the fetal liver. Although high levels of Runx1 expression have been reported in the early stages of myeloid differentiation, its expression decreases beyond the promyelocytic stage. Runx1 has been implicated in regulating a number of genes expressed early in the myeloid development pathway, including GM-CSF, macrophage colony-stimulating factor (M-CSF) receptor, MPO, NE, and IL-3. In addition to activating lineage-specific myeloid markers, Runx1 has been shown to stimulate the G 1 to S transition in myeloid and lymphoid cell lines.
A significant percentage (10% to 20%) of human leukemia has been found to be associated with mutations in the RUNX1 gene (see Chapters 59 and 60 ). The most common is the t(8;21) translocation, which generates the fusion protein RUNX1/RUNX1T1 (previously known as AML1-ETO), where the Runt domain of Runx1 is fused in frame with the RUNX1T1 (ETO) transcriptional corepressor. This fusion protein is thought to function predominantly as a repressor that inhibits expression of genes normally activated by Runx1. For example, the tumor suppressor p14 ARF , a critical Runx1 target encoded from the CDKN2A gene and which is necessary for p53 activation, is normally activated by Runx1 but is repressed by RUNX1-RUNX1T1. The mechanisms underlying Runx1 function through its target genes are not yet fully understood. However, studies in sea urchins have suggested that Runx1 regulates genes that contribute to chromatin architecture during cell proliferation. Runx1 functions within a narrow window during development by assisting in the opening of chromatin associated with genes that are vital to hematopoietic development, and for the formation of transcription factor complexes on these genes.
Studies involving mouse knockin models of Runx1-RUNX1T1 expression have demonstrated that the fusion protein alone is not sufficient to cause leukemia. However, these animals are more susceptible to mutagen-induced AML, suggesting that Runx1-RUNX1T1 is part of a multistep process that contributes to leukemogenesis.
C/EBPs are a family of basic region-leucine zipper (b-ZIP) transcription factors that recognize the consensus DNA-binding sequence 5′TKN NGYAAK3′ (Y = C or T; K = T or G) within the regulatory regions of target genes. C/EBP family proteins bind DNA as either homodimers or heterodimers. This family of transcription factors, which plays a crucial role in hematopoiesis, includes C/EBPα, -β, -γ, -δ, -ε, and -ζ (CHOP-GADD 153). These proteins share highly homologous C-terminal leucine zipper dimerization domains and positively charged basic region DNA-binding motifs but differ in their N-terminal transactivation domains. CHOP-GADD 153 lacks this domain altogether and instead dimerizes with and inhibits transactivation by C/EBPα, -β, and -ε.
With the exception of C/EBPε, which is expressed exclusively in the late stages of granulopoiesis and in T lymphocytes, the C/EBP family members are expressed in a wide variety of tissues, including liver, adipose tissue, lung, intestine, adrenal gland, peripheral blood mononuclear cells, and placenta. Both C/EBPβ and C/EBPδ are expressed at high levels in late-stage granulocytes. The C/EBP family members are known to exert pleiotropic effects in the tissues in which they are expressed. This may be because of their tissue- and stage-specific expression, their ability to dimerize with members of their own family and of the Fos/Jun and ATF/CREB families of transcription factors, and their ability to interact with other transcription factors such as nuclear factor-κB (NF-κB) and specificity protein-1 (Sp-1).
The C/EBP factors have been implicated in regulating differentiation in a variety of tissues. C/EBPα plays a role in adipocyte differentiation: inhibition of C/EBPα blocks adipocyte differentiation, and overexpression of C/EBPα induces adipocyte differentiation. Regulation of constitutive hepatic genes as well as acute phase response genes in the liver involves several C/EBP family members, in particular C/EBPα. Modulation of myelomonocytic differentiation is also attributed to the activity of C/EBP family members. The importance of this family of transcription factors in myeloid differentiation has been further demonstrated by the study of hematopoietic abnormalities observed in mice with targeted disruption of C/EBPα, -β, and -ε.
C/EBPα is thought to be a master regulator of the granulopoietic developmental program. It is expressed at high levels throughout myeloid differentiation and has been shown to bind to the promoters of multiple myeloid-specific gene promoters regulating gene expression at many different stages of myeloid maturation. Although Cebpa −/− mice die perinatally because of defects in gluconeogenesis that result in fatal hypoglycemia, they also have a selective early block in the differentiation of granulocytes without affecting either monocyte/macrophage maturation or the differentiation of other hematopoietic lineages. Myeloid cells of Cebpa −/− mice lack the G-CSFR, and it is thought that lack of mature neutrophils in these mice occurs as a result. However, the myeloid defect in Cebpa −/− mice is more severe than that seen in G-CSFR knockout mice, suggesting that C/EBPα has additional functions vital to granulocytic maturation.
CEBPA is a single exon gene, but it is expressed as two isoforms that arise from alternate translation start sites giving rise to a full-length C/EBPα p42 and a truncated dominant negative C/EBPα p30 isoform. Translational control of C/EBPα isoform expression is orchestrated by a conserved upstream open reading frame (uORF) in the 5′ untranslated region (UTR). This region is thought to be responsive to the activities of the translation initiation factors eIF4E and eIF2 (reviewed by Khanna-Gupta ) such that an increase in the activity of eIF4E or eIF2 results in an increase in expression of the shorter p30 isoform (reviewed by Calkhoven et al. ).
Several groups have reported loss of function mutations in the CEBPA gene in a subset of patients (~10%) with normal-karyotype AML (see Chapters 59 and 60 ) (reviewed by Muller and Pabst ). These mutations can be broadly classified into two categories. The first includes in-frame mutations clustered in the highly conserved C-terminus of the C/EBPα protein. The second category involves frameshift mutations at the N-terminus of C/EBPα resulting in the premature termination of the full-length C/EBPα p42 isoform while keeping the truncated C/EBPα p30 protein intact. The remaining C/EBPα p42 is thought to be rendered inactive by the dominant negative activity of the p30 isoform, although the mechanism is unknown. Mice that express a vector inducing overexpression of C/EBPα p30 from the Cebpa locus develop AML with complete penetrance. Thus changes in the expression ratio of the two C/EBPα isoforms play a role in cell fate and in leukemogenesis (reviewed by Muller and Pabst and Kirstetter et al. ).
The expression of C/EBPα is associated with growth arrest and differentiation of granulocyte precursor cells. This block in proliferation is thought to occur via the interaction of C/EBPα with, and inhibition of, the cyclin-dependent protein kinases cdk2 and cdk4. In addition, C/EBPα inhibits E2F-dependent transcription, which in turn leads to inhibition of cell proliferation and induction of differentiation associated with C/EBPα-induced granulopoiesis.
Expression of C/EBPβ increases during myeloid maturation and is important for monocyte/macrophage gene expression and development. Mice lacking the Cebpb gene demonstrate reduced B-cell numbers and defects in macrophage activation and function and increased susceptibility to microbial infections. Knockout studies reveal that C/EBPβ is not essential for myeloid development, although knockin of Cebpb into the Cebpa locus of Cebpa −/− mice rescues granulopoiesis. Several monocyte/macrophage-specific genes are activated by C/EBPβ, including G-CSFR, lysozyme, CD11c, monocyte chemoattractant protein-1 (MCP-1), IL-6, IL-8, and nitric oxide synthase. As with C/EBPα, multiple isoforms of C/EBPβ are generated from a single transcript through three alternate translation initiation sites and a leaky ribosome scanning mechanism. The shortest of these isoforms, initiated at the most distal start codon, results in the formation of liver-enriched inhibitory protein (LIP), which lacks the N-terminal activation domain present in full-length C/EBPβ and has been implicated as a negative regulator of C/EBPβ function. It has been suggested that the ratio of C/EBPβ to LIP may affect cellular proliferation and differentiation. The activity of C/EBPβ is regulated through protein-protein interactions and posttranslational modifications. For example, in early myeloid progenitor cells, C/EBPβ is found in an unphosphorylated state in the cytoplasm, whereas on differentiation, C/EBPβ becomes phosphorylated and translocates to the nucleus.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here