Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Current standards for prenatal genetic screening and testing are highly focused on detection of aneuploidies that are compatible with live birth, including trisomy 21, which affects 1:600 newborns overall and is associated with long-term survival, and the less common and more severe trisomies 18 and 13, which respectively affect 1:5000 and 1:15,000 liveborn infants. When an amniocentesis or other prenatal diagnostic procedure is performed for fetal structural abnormalities detected by fetal imaging (ultrasound and/or MRI), aneuploidy is found in fewer than 1% to 30%–40% of cases, depending on the type of anomalies and the presence of single versus multiple anomalies. The addition of chromosomal microarray analysis (CMA) provides an incremental detection rate of clinically significant copy number variants in 6%–7% of fetuses with prenatally diagnosed congenital anomalies and of more than 10% when there are multiple fetal anomalies. When amniocentesis or chorionic villus sampling (CVS) is performed for other indications, such as advanced maternal age or positive aneuploidy screening, the incremental diagnostic yield of CMA is 1%–1.7%. Although CMA therefore offers significant diagnostic improvements over karyotype analysis, this standard workup still leaves approximately 60% or more pregnancies with fetal anomalies without a genetic diagnosis, in large part because testing does not include single gene disorders. Although single gene disorders are individually rare, their collective disease burden is highly significant with an estimated frequency of 0.36% of live births or 1% overall. There are currently >6000 single gene disorders and traits caused by pathogenic variants in >3800 genes listed in the Online Mendelian Inheritance of Men (OMIM) database ( https://omim.org/statistics/geneMap accessed April 3, 2018). Overall, autosomal recessive genetic disorders account for 6%–8% of pediatric hospital admissions, in contrast to only 0.4%–2.5% for chromosomal abnormalities. Single gene disorders are also responsible for 20% of infant mortality. Despite these numbers, these conditions are not included in traditional prenatal testing paradigms, or in guidelines for prenatal genetic workup for fetuses with congenital anomalies, largely because until recently it was difficult and impractical to determine which specific gene(s) to assess. With the development of high-throughput next-generation sequencing (NGS) technology, an entire genome or exome can now be sequenced in days. This has simultaneously accelerated the pace of new disease gene discovery and the development and diagnostic use of multigene panels, as well as whole exome sequencing (WES), which analyzes the 1%–2% of the genome that codes for expressed RNAs and proteins, and whole genome sequencing (WGS). Diagnostic WES is being increasingly integrated into pediatric and adult genetic disease evaluation, where the incremental detection rates of disease-causing variants ranges from 25% to more than 50%, depending on the clinical indication. We review here WES technology, its applications, and the still limited experience with its use for prenatal genetic diagnosis. Although early data on the utility of fetal WES for prenatal diagnosis are promising and suggest its value, they also highlight some pitfalls that arise from detection of variants of uncertain significance (VUSs), secondary or incidental findings, and unanticipated diagnoses in family members. These can all result in complex pre- and posttest counseling situations.
For NGS, genomic DNA is first sheared into small fragments of about 50 to up to a few hundred nucleotides, and adapters are linked to one or both ends of these fragments. In WGS, where the entire genome is sequenced, this pool of fragments is called the sequencing library ( Fig. 13.1 ). In contrast, with WES, only the 1%–2% of the genome that contains the coding exons, which harbor up to 85% of all known disease-causing mutations, is sequenced. With WES, the DNA fragments that overlap with exons and their flanking introns are first purified from the entire library by hybridization to a collection of DNA baits of known sequence that are attached to magnetic beads to generate an enriched sequencing library. A similar enrichment strategy is used for high-throughput sequencing with disease-specific multigene panels. The obtained sequencing library is then immobilized on a solid surface, amplified in clusters, denatured, and then sequenced by synthesis of a new complementary strand. In this process, each nucleotide (A, C, G, T) that is incorporated has a different fluorescent tag, so that its insertion at a specific location in the sequenced fragment can be recorded. This is repeated multiple times in “massively parallel sequencing” for each nucleotide present in the overlapping fragments, resulting in rapid, accurate identification of the incorporated nucleotide for each cluster. The obtained sequence reads are then aligned using bioinformatics tools to generate a consensus sequence that is compared with the human reference sequence.
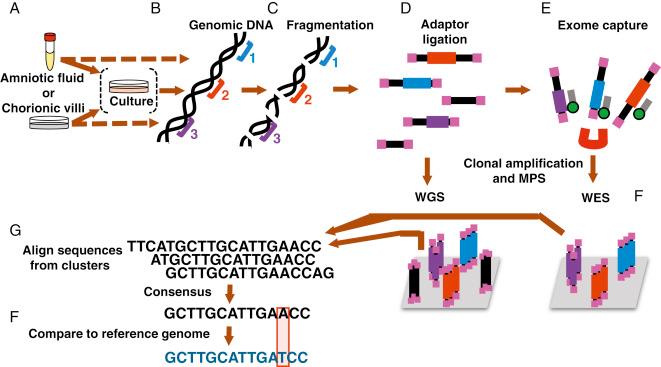
The quality of the obtained sequence varies, and two parameters used to describe the quality are the sequencing depth, which refers to the number of overlapping reads for each base pair, and the sequence coverage, which refers to the fraction of the sequence that is covered at sufficient depth. The American College of Medical Genetics and Genomics (ACMG) recommends that for diagnostic WES, ≥ 90%–95% of the sequence should be covered at least 10-fold and that the average depth should be ≥100-fold. (see Fig. 13.1 ).
Compared with its use for children or adults, genome-wide sequencing for prenatal diagnosis presents unique technical challenges. Prenatally obtained samples, usually amniotic fluid or chorionic villus samples (CVS), are often smaller in volume with lower numbers of nucleated cells and may require a cell culture step before enough DNA can be made to prepare a sequencing library. Because the sample is obtained through an invasive procedure, there is also a small risk for contamination with maternal cells. In prenatal diagnosis, timing of the diagnostic test and rapid turnaround times are critical if the goal is to have results that can inform prenatal, perinatal, and postnatal management. Laboratories that offer prenatal genome-wide sequencing-based tests must therefore implement procedures to assure rapid turnaround times from sample submission to return of results. This is achieved by a combination of improvements of sequencing equipment, reagents and protocols, use of optimized workflows, including a trio-sequencing approach, wherein the fetal sample and parental samples are sequenced and analyzed in parallel to aid interpretation (see below).
Each sequenced human genome is unique and contains hundreds of thousands of sequence variations when compared with the reference genome, but the vast majority of these are benign and not associated with disease. Although there are fewer variants, in the range of 20,000–50,000, identified with WES, determining which of those are responsible for disease or a developmental phenotype is a highly complex process.
Diagnostic and research laboratories typically analyze sequence data in a stepwise approach using a combination of strategies, including comparison with databases of sequence variants found in healthy individuals and in various pathological states, together with bioinformatics tools that predict the functional impact of specific variants on the gene and its encoded protein. This allows filtering out of benign variants and prioritizing those that are most likely to be causative for the developmental phenotype or disease for which the sequencing was performed. In the United States, diagnostic laboratories offering WES abide by standards of variant interpretation and reporting developed by organizations such as the ClinGen Clinical Genome Resource ( https://www.clinicalgenome.org/ ) and the ACMG to classify variants into five categories: pathogenic, likely pathogenic, VUS, likely benign, and benign.
The commonly used databases of genomic variants that are used to guide interpretation include the exome aggregation consortium (ExAc) database, genome aggregation database (gnomAD), clinical variation database (ClinVar), Human Gene Mutation Database (HGMD), and Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources (DECIPHER). Sequence variants that are common in healthy individuals (single-nucleotide polymorphisms; SNPs) are more likely to be benign, whereas unique rare variants are more likely to be pathogenic and need to be further interpreted. Reports of other individuals with identical or other pathogenic variants in the same gene who have a known disorder or an overlapping phenotype strongly support pathogenicity, although there can be exceptions because of incomplete penetrance, variable expressivity, or misclassification of variants.
The functional consequence of a variant can also be predicted using bioinformatics tools such as PolyPhen 2 ( http://genetics.bwh.harvard.edu/pph2/ ), which classifies variants as probably damaging, possibly damaging, or benign ; SIFT (Sorting Intolerant from Tolerant; http://sift.jcvi.org/ ), which provides a score that indicates whether an amino acid change is predicted to be tolerated or damaging ; or MutationTaster2, which predicts if a sequence alteration is likely to be disease-causing or a benign polymorphism. Typically, but not always, variants such as stop-gain (nonsense) or frameshift mutations that are predicted to lead to a complete absence of the produced protein are more likely to be pathogenic, whereas missense variants, which result in the substitution of one amino acid for another, can have variable effects.
The inheritance pattern of a rare variant in the family is also an important consideration. In inherited autosomal dominant disorders, the pathogenic or likely pathogenic variant is typically heterozygous and segregates in a family’s pedigree with the inherited disease, but a “de novo” dominant variant is only found in the affected individual and not in either parent. For autosomal recessive disorders, the affected individual has a pathogenic or likely pathogenic variant on each allele of a gene, usually inherited “in trans” from each of the biological parents, who are carriers. Variants on the X chromosome cause X-linked inherited or de novo conditions. At times, supporting information from animal models or functional assays can be used to further refine the interpretation.
The final step, clinical interpretation, is critically important and requires collaboration and information sharing between the clinician and the diagnostic laboratory.
These combined approaches for variant interpretation rely greatly on publicly available data. Thus, because WES and WGS are emerging diagnostic and research tools for identifying the causes of birth defects and rare genetic disorders, data sharing in a manner that safeguards individual privacy has been formally recommended by professional societies and is critical for advancing our understanding of pathogenicity and ability to most optimally apply WES or WGS for prenatal diagnosis.
Although the databases, tools, and strategies outlined above are standardized for interpretation of any sequencing results, there are unique challenges in the prenatal setting that must be considered. All currently used databases are primarily populated with variant data originating from pediatric and adult presentations of genetic diseases and include very limited data from prenatal or fetal phenotypes. This is particularly challenging for prenatally lethal conditions that are not reported in neonates or children, as they may not survive to birth. An important aspect of WES interpretation is correlation of a discovered rare variant with an already known clinical phenotype that matches the phenotype of the sequenced proband. However, the proband’s phenotype is often incompletely ascertained prenatally because of imaging limitations or because some features are not present until after birth or later in life. Early data from prenatal WES use have also revealed new unpredicted prenatal findings associated with known disease genes, resulting in “phenotypic expansion” of the features of known single gene disorders to include previously unascertained prenatal phenotypes.
An important current initiative in clinical genetics is the use of a standard phenotyping nomenclature, by using Human Phenotype Ontology (HPO) terms, such that there is uniformity in phenotype descriptions in shared clinical data associated with specific variants from different centers. Like other databases, HPO terms also contain relatively limited clinically useful prenatal phenotypic descriptions. Initiatives are underway to include more prenatal phenotypes in clinical databases.
Because WES and WGS are genome-wide tests, pathogenic variants in genes that cause single gene disorders unrelated to the indication for prenatal testing can be found incidentally. If the identified single gene disorder usually presents after birth, in childhood, or even in adulthood, a fetus with such a “genotype first” result will not have clinical features of the disease until later in life. How to manage such findings is particularly challenging when the variant is a VUS or causes an adult-onset disorder. Laboratories and clinicians must therefore develop strategies for reporting such variants and informing parents about them.
Although NGS is a powerful tool in evaluating the human genome, this technology has limitations in which types of variants can be detected ( Table 13.1 ). It is well suited for detecting sequence variants that affect one or a few nucleotides (i.e., single-nucleotide variants; SNVs) in unique gene sequences, including missense mutations that result in a change in amino acid in the encoded protein; nonsense and frameshift mutations that cause premature stop codons (stop-gain mutations) resulting mostly in the absence of the protein; and splice-site mutations, which can disrupt exon-splicing, thereby altering the encoded protein. However, the final assembled sequence in NGS relies on the alignment of overlapping relatively short sequenced fragments, which does not work well for sequences with high homology to another sequence in the genome, such as duplicated genes or exons; pseudogenes; and highly homologous gene families. Repeat sequences, including endogenous repeats and triplet repeat amplification mutations, such as the CGG repeat expansion in the 5′ untranslated region of the FMR1 gene that causes fragile X syndrome, are also difficult to detect by NGS. In addition, detection of structural chromosomal abnormalities or aneuploidy from WES is challenging and not currently feasible for clinical use; CMA remains the method of choice for clinical diagnosis of larger unbalanced chromosomal abnormalities (see Chapter 12 ). In contrast, larger-size copy number changes, structural chromosomal abnormalities, and aneuploidy can be detected by low-coverage WGS, which can be used for detection of aneuploidy and for prenatal and preimplantation diagnosis. Many of the currently used noninvasive cell-free DNA screening strategies for fetal aneuploidy rely on the principle of counting fragments sequenced through low-coverage WGS of maternal plasma cfDNA. (see Chapter 9 ).
| Variant Type | NGS Method | ||
|---|---|---|---|
| Multigene Panel | Whole Exome Sequencing | Whole Genome Sequencing | |
| Aneuploidy | – | – | ++ |
| Balanced chromosome rearrangements | – | – | + |
| Large CNV: >5 Mb | – | – | + |
| Intermediate CNV: 1–5 Mb | – | – | ++ |
| Small CNV: 0.1–1 Mb | ++ | + | ++ |
| Insertions/deletions <100 bp | +++ | +++ | +++ |
| SNV in unique exons | +++ | +++ | +++ |
| SNV at intron/exon boundary | +++ | +++ | +++ |
| SNV in introns | – | – | +++ |
| SNV in region of high homology | ? | – | – |
| Mosaic SNV >15–20% a | ++ a | ++ a | ++ a |
| Trinucleotide repeat expansion | – | – | – |
a Algorithms for interpretation may need adjustment to detect mosaicism.
WES focuses on the 1%–2% of the genome that includes the coding sequences, whereas WGS has the potential to provide information on variants in other important regions, such as promoters and regulatory elements. For this reason, WGS has a higher diagnostic capacity, although it is more complex to confirm that a detected sequence variant truly causes the condition for which the sequencing was done, and often requires functional studies in cell lines or animal models. This is an important reason, in addition to higher cost, why diagnostic WGS is not currently routinely used by most laboratories.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here