Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
It has been known for decades that genetic alterations are a fundamental driving force in the initiation and progression of human cancers. It is also now apparent, through a more recent and growing body of work, that epigenetic changes may be equally important in tumor development. Epigenetics refers to heritable changes in gene expression in somatic cells that are determined by other than alterations in the primary base sequence of DNA. In essence, the primary sequence of DNA provides the “hard drive,” storing all information to determine cell phenotypes. However, to selectively function, DNA requires the “software packaging” of epigenetics to guide patterns of heritable gene expression. In this way, different cell phenotypes emerge against a common DNA sequence background to facilitate processes such as embryonic development and cell differentiation. Just as DNA mutations can mediate individual stages of tumor development by fostering over-, or under-, function of key genes, epigenetic abnormalities can heritably allow similar gene expression aberrations. In this chapter, features are outlined of this latter form of altered gene function and how it is coming to affect the understanding and management of human cancer.
To specify regions of DNA that contribute to gene expression patterns in a cell, the exposure of sequences to, and the function of, the transcriptional machinery is controlled by packaging of the DNA in the nucleus. This is accomplished, as described later, through a complex and dynamic interaction of DNA with proteins to constitute cellular chromatin, which undergoes posttranslational modifications to establish the transcriptional and regulatory activity of DNA regions and individual genes. This DNA regulation is also modulated by patterns of the only postreplicative modification made directly to DNA, methylation at cytosines located ahead of guanosines (the CpG dinucleotide) in the genome. Alterations to all of these processes are being increasingly identified as important to the evolution of cancer.
The basic molecular unit of DNA packaging, the nucleosome is a structure characterized by the wrapping of approximately 146 bp of DNA around what is termed the histone octamer consisting of a tetramer of two histone H3, H4 dimers and a dimer of histone H2. Groups of nucleosomes may in turn be organized into higher order structures through the actions of chromatin remodeling complexes. Tightly compacted nucleosome aggregates, or “closed chromatin,” are characteristic of DNA regions that are transcriptionally silent, versus more irregularly and linearly spaced nucleosomes, or “open” chromatin, characteristic of DNA regions where transcription is active.
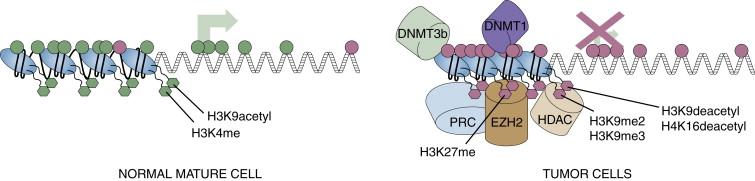
One of the most exciting and dynamic areas of chromatin biology concerns another key facet of nucleosome function, posttranslational modification of key amino acid residues of the histones, that helps determine the transcriptional status of genomic regions ( Figure 5-1 ). These modifications constitute what has been termed the histone code for gene expression regulation. Thus, acetylation of key residues, such as lysine 9 of histone H3 (H3K9acetyl), by enzymes known as histone acetylases (HATs) usually specifies for transcriptionally active regions while deacetylation at these sites, mediated by histone deacetylases (HDACs), is usually associated with transcriptional repression. Methylation of key amino acids also occurs and may be an activating or inactivating mark, depending on the site. For example, the mark of H3K4 methylation is enriched at active areas, whereas H3K9 methylation or H3K27 methylation is characteristic of transcriptionally repressed regions. These methylation marks are controlled by a dynamic process involving individual histone methyltransferases, which place the marks, and demethylases, which can remove them.
Interacting with all of the dynamics for nucleosome assembly, placement, and histone modifications, to mediate nuclear packaging of DNA, is the modification of DNA methylation (see Figures 5-1 and 5-2 ). This process, which as noted earlier occurs predominantly in all but embryonic stem cells (ESCs) at CpG dinucleotides, is mediated by three DNA methyltransferase (DNMT) enzymes that utilize S -adenosylmethionine as a methyl donor group to transfer this moiety for covalent linkage to the cytosines. DNA methylation adds a dimension to packaging of DNA and nucleosomes into repressive domains by stabilizing the heritable nature of transcriptional silencing. A key aspect of this dimension concerns the distribution of DNA methylation in the genome (see Figure 5-2 ). In most genomic regions, the CpG dinucleotide is underrepresented because, over evolution, these cytosines have been depleted through deamination of methylcytosines to form thymines. However, as many as 80% of these remaining CpG sites are DNA methylated in the human genome, and this has an important functional correlate. This methylation corresponds to the fact that most of our genome, in adult cells, is packaged away into nucleosome-compacted DNA characteristic of regions of transcriptional repression (see Figure 5-2 ). This may constitute one of the most important functions of genomic DNA methylation, which is to ensure tight heritability of overall genomic transcriptional repression to prevent unwanted expression of elements such as viral insertions, repeat elements, and other potentially deleterious sequences.
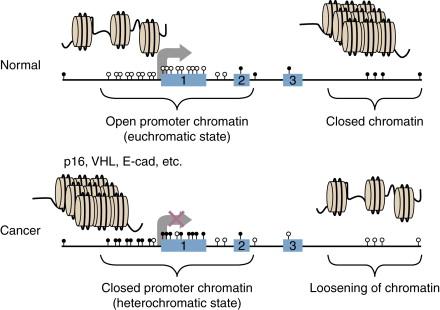
In contradistinction to the depletion of CpGs throughout most of the genome, approximately half of the genes in the genome have regions in their promoters, termed CpG islands where the expected frequency of the nucleotide has been preserved (see Figure 5-2 ). For most such genes, these islands are protected from DNA methylation, and this methylation-free state is associated with active transcription of these genes, or preservation of their being in a transcription-ready state. These CpG islands are the target of key epigenetic abnormalities in cancer cells, as discussed in detail in subsequent sections of this chapter.
In addition to the previously described role of DNA methylation in global DNA packaging, it is also linked to regulation of expression for specific genes in normal cells. In this regard, when localized to gene promoter regions, it may act to provide a tightening of heritable states for gene silencing. Examples include the imposition of DNA methylation in the promoters of genes shortly after other processes initiate their silencing in regions on the inactive X-chromosome of females. A similar role is apparent in genes that are imprinted in mammals wherein DNA methylation of promoter regions is seen on the silenced allele of such genes. DNA methylation also may participate in regulating expression of certain genes in normal cells that are expressed in a tissue-specific manner, such as the silencing of globin genes in all but cells actively engaged in erythropoiesis.
In the gene-silencing roles, there is a tight interplay between the modification of key histone amino acid residues and DNA methylation. Thus, at least in lower organisms such as Neurospora and Arabidopsis, methylation of lysine 9 of histone H3 (H3K9me) may help determine positions where cytosine methylation is placed in the genome. Increased levels of the active histone modification H3K4 methylation can be inhibitory to the recruitment of the DNA methylating enzymes. In turn, DNA methylation recruits a series of proteins, methylcytosine binding proteins (MBPs), which are complexed, in turn, with HDACs, which help maintain the deacetylation of H3K9 and other key histone lysines in regions of silenced genes.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here