Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
DNA repair is central to the field of cancer biology, and it has important implications for cancer diagnosis and treatment. Cancer cells are often deficient in a normal DNA repair function, and this deficiency allows the tumor to develop genomic instability. With defective DNA repair, the tumor cell can break and reform chromosomes, generate new oncogenic fusion genes, disrupt tumor suppressor genes, amplify drug resistance genes, and progress to a more malignant state. A DNA repair deficiency also accounts for the enhanced sensitivity of tumor cells to genotoxic agents, such as ionizing radiation (IR) and genotoxic chemotherapy. A specific defect in homologous recombination DNA repair also renders a tumor cell hypersensitive to polyADP ribose polymerase (PARP) inhibitors. A thorough knowledge of DNA repair mechanisms in normal and cancer cells may therefore lead to better clinical management of cancer.
In order to understand the process of DNA repair, one must first consider the wide range of DNA-damaging events in a cell. DNA may undergo spontaneous damage, such as deamination of cytosine or spontaneous hydrolysis of the phosphodiester backbone. DNA may develop mismatched bases, perhaps resulting from the deployment of an error-prone DNA polymerase during S phase progression. DNA may be attacked by reactive oxygen species (ROS). Indeed, some of the most sophisticated DNA repair mechanisms in a cell are mechanisms that cope with the removal of oxidative DNA lesions.
Of particular relevance to cancer is the DNA damage from alkylating agents or from ultraviolet (UV) light or IR. DNA damage resulting from these environmental agents can lead to heightened mutagenesis and oncogenesis. Also, many of these agents themselves have anticancer activity. Thus, DNA-damaging agents can cause human cancer but, ironically, are among the primary means available to clinicians for treating cancer. Accordingly, some chemotherapeutic agents have effective anticancer activity in the short run but are responsible for causing secondary cancers in the long run.
Most anticancer agents function by directly damaging DNA. Effective anticancer drugs include monofunctional alkylating agents (cyclophosphamide, BCNU), bifunctional alkylating agents, (cisplatin, carboplatin, oxaliplatin), and DNA intercalating agents (adriamycin). In addition, IR and the radiomimetic agent bleomycin can cause double-strand breaks in DNA directly. Bleomycin is a small glycopeptide that chelates ferrous ion and binds to specific sequences of double-stranded DNA containing pyrimidine repeats. In the presence of oxygen, bleomycin generates a local high concentration of hydroxyl radicals capable of causing local double-strand breaks. Other drugs, such as the topoisomerase inhibitors etoposide and camptothecin, can lead to the accumulation of DNA damage. Thus, known anticancer agents can generate a wide range of DNA damage, including damaged bases, single-strand breaks, and double-strand breaks.
It is important for oncologists to bear in mind that anticancer drugs generate their cytotoxic effects through DNA damage. First, effective anticancer protocols often include a combination of chemotherapeutic agents and IR. Together, these agents cause a broader spectrum of DNA damage in the tumor than single agent therapy (monotherapy). This broad spectrum may contribute to the synergy observed with these agents. Second, chemotherapy combinations are often chosen to limit toxicity to normal tissue. Agents that generate the same class of DNA damage (such as IR and bleomycin, which both generate double-strand breaks) may have enhanced toxicity compared to other combinations. Some newer classes of drugs inhibit normal DNA repair processes. These so-called chemosensitizers may be particularly effective when used in combination with a more traditional cytotoxic, DNA-damaging drug. A combination of a DNA repair inhibitor and a direct DNA damaging agent can also result in significant toxicity to normal tissue. One way to limit this toxicity would be to deliver one agent such as the chemosensitizer systemically but to deliver the other agent, such as IR, locally to the tumor.
DNA repair is strictly defined as the cellular responses that are associated with the restoration of the normal base pair sequence and structure of damaged DNA. As described in the following section, there are six primary DNA repair pathways, and each pathway is composed of a series of biochemical events leading to the sensing, excision, and restoration of the normal DNA sequence.
It is instructive to consider the history of DNA repair research as it relates to cancer biology. Early studies of DNA repair evolved from the study of normal DNA replication and metabolism. These early studies relied heavily on the use of damaged DNA templates as substrates for the purification of DNA repair enzymes. Such templates were incubated with cell-free extracts, and the recovered DNA was analyzed for specific incision and excision events. Not surprisingly, these assays uncovered many of the pertinent endonuclease and exonuclease activities required for DNA repair. It has become increasingly apparent that DNA repair proteins are assembled in protein/protein complexes, such as the excision repair complex or the mismatch repair complex. Still, the regulatory networks and relevant posttranslational modifications of DNA repair proteins (i.e., phosphorylations and ubiquitinations) were largely missed by these early biochemical studies.
The study of inherited human DNA repair disorders also contributed greatly to the recognition of the six major DNA repair pathways (see later discussion). These studies depended on the establishment of mutant human cell lines derived from patients with genetic diseases. For instance, in 1968, James Cleaver isolated fibroblast lines from humans with the disease xeroderma pigmentosum (XP). Importantly, these lines retained their UV light–hypersensitivity phenotype and have been invaluable tools for somatic cell fusion, complementation analysis, and expression cloning of XP genes. Subsequently, other investigators were able to establish mutant cell lines from humans with other DNA repair disorders such as ataxia-telangiectasis (A-T), Fanconi anemia (FA), and Nijmegen breakage syndrome (NBS). These cell lines continue to be used extensively as models of human cancers that also lack the relevant DNA repair pathways.
The study of model organisms has contributed greatly to our understanding of DNA repair processes. For instance, investigators isolated mutants in the yeast Saccharomyces cerevisia e, which were hypersensitive to UV light, IR, or DNA crosslinking agents. In many cases, the genes that were mutated in these yeast strains cooperated in common DNA repair and DNA damage response pathways. IR-induced double-strand breaks in DNA are normally repaired by the DNA repair process of homologous recombination (HR). Accordingly, many of the relevant genes corresponding to these mutant strains and required for normal HR repair were first isolated in yeast. Thereafter, the human homologues of these genes were identified. Other model organisms and cell lines have been especially important in the identification of genes involved in DNA repair pathways, such as mismatch repair and translesion DNA synthesis. Among the most useful model systems for studying DNA repair are the Caenorhabditis elegans and chicken (DT40) genetic systems.
In the postgenomic era, and following the identification of a large number (perhaps 130) of distinct DNA repair proteins, investigators have turned to x-ray crystallography for a detailed understanding of DNA repair protein interaction with damaged DNA. The structures of many endonucleases, helicases, and ligases are now available, providing the opportunity for computer-assisted drug development (CADD) of DNA repair enzyme inhibitors. Also, mass spectrometry has been used to identify critical posttranslational modifications of DNA repair proteins. These modifications appear to be essential to the proper localization and assembly of DNA repair complexes around sites of DNA damage. The modifications may also regulate the intrinsic catalytic activity of the repair complexes. These protein modifications can be used as surrogate markers, or biomarkers, of DNA repair activity in a given tumor type as well (see later discussion).
As described previously, the combination of (1) biochemistry with damaged DNA templates, (2) human mutant cell lines with genetic deficiencies of DNA repair, (3) genetics of yeast mutants with IR or UV sensitivity, and (4) structural studies of DNA repair proteins has led to the establishment of six major DNA repair pathways. These pathways are base-excision repair (BER), nucleotide-excision repair (NER), mismatch repair (MMR), homologous recombination (HR), nonhomologous end joining (NHEJ), and translesion DNA synthesis (TLS) ( Table 4-1 ).
| DNA Damage Repair Pathway | Function | Examples of Gene Mutation | Examples of Altered Expression of a Normal Gene | Effect of Loss of Pathway on Clinical Response |
|---|---|---|---|---|
| Base-excision repair (BER) | Repair of damaged bases or single-strand DNA breaks | None reported | None reported | None reported |
| Mismatch repair (MMR) | Repair of mispaired nucleotides | Mutation of MSH2, MSH6, and MLH1 in Turcot syndrome (brain and colon tumors) and HNPCC (colon and gynecologic cancers) | Loss of expression of MSH2 or MLH1 in sporadic colon cancer | Resistance to DNA monoadducts Sensitivity to DNA crosslinks |
| Nucleotide-excision repair (NER) | Excision of a variety of helix-distorting DNA lesions | Mutation of XPA, XPB, XPC, XPE, XPF, or XPG in xeroderma pigmentosum (skin cancer) Variant expression of ERCC1 or XPD in lung cancer |
Loss of XPA expression in testicular germ-cell tumors | Sensitivity to DNA adducts |
| Homologous recombination (HR) | Repair of double-strand DNA breaks | BRCA1/2 mutated in early-onset breast/ovarian, prostate, pancreas, and gastric cancers FANC genes mutated in Fanconi anemia |
Loss of expression of BRCA1/2 in ovarian and lung cancers Loss of NBS1 expression in prostate cancer |
Sensitivity to DNA double-strand breaks |
| Nonhomologous end joining (NHEJ) | Repair of double-strand DNA breaks | DNA ligase IV mutated in Lig4 syndrome (leukemia) Artemis mutated in Omenn syndrome (lymphoma) |
Loss of Ku70 expression in cervical, rectal, and colon cancers Loss of Ku86 expression in rectal cancer |
Sensitivity to DNA double-strand breaks |
| Translesional synthesis (TLS) | Bypass of DNA adducts during DNA replication | DNA pol E mutated in xeroderma pigmentosum variant (XPV; skin cancers) | Pol β overexpressed in uterus, ovary, prostate, and stomach cancers Pol iota overexpressed in breast cancer |
Resistance to DNA adducts |
There is also considerable redundancy in the function of the DNA repair pathways. When one pathway is disrupted, another pathway can partially compensate, especially if the second pathway is upregulated. For instance, a cell that is deficient in HR repair may depend more on the error-prone NHEJ repair pathway for the repair of double-strand breaks. Also, thymine dimers, which are generated by UV light exposure, can be repaired by NER repair or bypassed and effectively ignored by TLS polymerases. In some cases, the absence of one DNA repair pathway results in a hyperdependence on one or more other DNA repair pathways. This so-called synthetic lethality among DNA repair pathways has important implications for the design of new anticancer drugs (see later discussion).
The six DNA repair pathways are not constitutively activated, but instead are highly regulated. The pathways are often activated at discrete times in the cell cycle. For instance, HR repair and TLS repair are active during the S phase of the cell cycle. Also, the DNA repair pathways are differentially active in various tissues and cell types. For instance, HR and TLS are more active in rapidly growing cells, such as hematopoietic cells, whereas NHEJ is more active in postreplicative cells. Accordingly, absence of a particular DNA repair pathway may be particularly disruptive to the growth and survival of some normal tissues and some cancers. Here is a brief description of the six DNA repair pathways, with an emphasis on the enzymes in the pathways and the preference for DNA lesions repaired.
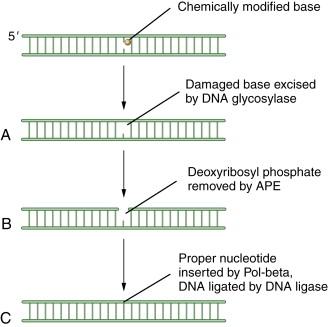
BER has been reviewed by Wilson. The cell uses BER to correct damaged DNA bases or single-strand DNA breaks. These lesions often result from spontaneous DNA damage (DNA deamination or hydroxylation of bases) or from exposure to environmental alkylating agents. In this pathway, damaged bases are removed by one of at least 10 DNA glycosylases. The resulting apurinic/apyrimidinic (AP) sites are processed first by the Ape1 AP endonuclease, leaving a 5′ deoxyribose phosphate; then by an AP lyase activity, leaving a 3′-elimination product. Single-strand breaks are then filled in by a DNA polymerase, either with a single nucleotide or with a longer repair patch, followed by ligation. The latter events in the BER pathway are regulated by the enzyme PARP1 (polyADP ribose polymerase 1). During the DNA damage response, the PARP1 enzyme polyADP ribosylates BER enzymes and enhances BER activity. Accordingly, tumors that have high levels of BER activity may be hypersensitive to PARP inhibitors. A schematic representation of BER is shown in Figure 4-1 .
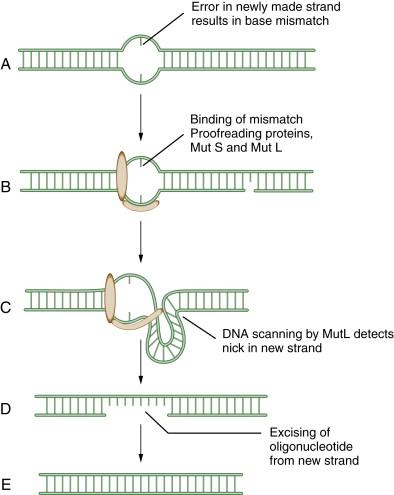
MMR has been reviewed by Modrich. MMR rapidly removes mispaired nucleotides that result from replication errors and is also involved in the detection and repair of DNA adducts such as those resulting from platinum-based chemotherapeutic agents. Initially, the heterodimeric MSH complex recognizes the nucleotide mismatch, followed by its interaction with MLH1/PMS2 and MLH1/MLH3 complexes. Several proteins participate in the process of nucleotide excision and resynthesis. Tumor cells deficient in MMR have much higher mutation frequencies than normal cells and exhibit microsatellite instability, a genomic biomarker of the underlying defect. Patients with the genetic disease HNPCC (hereditary nonpolyposis colon cancer) have germline mutations of MMR genes and are predisposed to MMR-deficient colon cancers. Recent studies suggest that MMR-deficient cells may be hypersensitive to inhibitors of various DNA polymerases, such as POLB and POLG. At least six genes—MSH2, MLH1, PMS2, MSH3, MSH6, and MLH3—are involved in mismatch repair. A schematic representation of MMR is shown in Figure 4-2 .
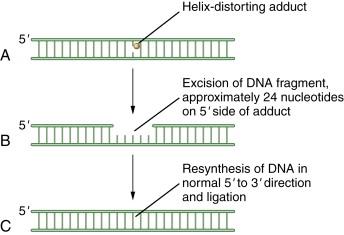
NER acts on a variety of helix-distorting DNA lesions, caused mostly by exogenous sources that interfere with normal base pairing. This pathway may be particularly important in the response to adduct-forming chemotherapeutic agents such as platinum-based chemotherapy. The primary function of NER appears to be the removal of damage such as pyrimidine dimers, which are induced by UV light. Members of the NER pathway include the XPA, XPB, XPC, XPD, XPE, and XPG proteins. Two other NER proteins, XPF and ERCC1, are especially important for the processing of DNA crosslink repair. Recent studies indicate that monitoring the levels of these proteins in tumors may provide important biomarkers for predicting crosslinker drug sensitivity. For instance, some non–small-cell lung cancers are deficient in ERCC1, and this deficiency correlates with the cisplatin sensitivity of the specific tumor.
As for the other DNA repair pathways, these proteins cooperate to recognize and excise the damaged nucleotides and resynthesize and ligate the damaged DNA strand. In the process of NER, initially a DNA-binding component, the DDB, binds to sites of damaged DNA, such as cyclopyrimidine dimers or 6-4 photoproducts. The DDB consists of DDB1 and DDB2. Mutations in the DDB2 gene cause the genetic complementation group XPE. DDB is part of a ubiquitin E3 ligase that polyubiquitinates XPC. Polyubiquitination of XPC enhances its DNA binding. This binding sets the stage for the downstream binding of the entire excision repair complex, TFIIH, thus leading to excision of the damaged bases.
Eukaryotic NER includes two major branches, transcription-coupled repair (TCR) and global genome repair (GGR). GGR is a slow, random process of inspecting the entire genome for injuries, whereas TCR is highly specific and efficient and concentrates on damage-blocking RNA polymerase II. The two mechanisms differ in substrate specificity and recognition, and hence the enzymes involved are important nodal points for posttranslational modifications. A schematic representation of NER is shown in Figure 4-3 .
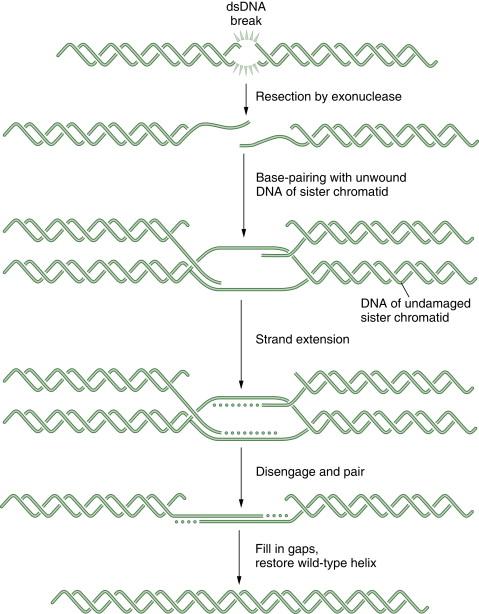
DNA double-strand breaks (DSBs) can be caused by many different environmental factors, including reactive oxygen species, IR, and certain antineoplastic drugs, such as bleomycin, anthracyclines, and topoisomerase inhibitors. Alternatively, DSBs can result from endogenous factors, especially during normal S-phase progression. Failure to repair DSBs can lead to a number of consequences, including mutations, gross chromosomal rearrangements, and other aberrations and eventually cell death. HR is a process by which DSBs are repaired through the alignment of homologous sequences of DNA and occurs primarily during the late S to M phase of the cell cycle. Initially the RAD50, MRE11, and NBS1 complex, which possesses a 3′-5′ exonuclease activity, exposes the 3′ ends on either side of the DSB, a process that may also require BRCA1. The 3′ advancing strand from the damaged chromosome then invades the complementary sequence of the homologous chromosome. The breast cancer susceptibility protein BRCA2 and the single-strand DNA binding protein RAD51 are required for the process. The 3′ end of this strand is then extended by an HR polymerase, by reading off of this complementary sequence. After replication has extended past the region of the DSB, the 3′ end of the advancing strand returns to the original chromosome, and replication continues. A schematic representation of HR is shown in Figure 4-4 . HR repair is especially important in the repair of DSBs and DNA interstrand crosslinks. Because some tumors, particularly breast and ovarian tumors, are defective in HR repair, drugs that cause these lesions may be particularly effective in this setting.
NHEJ, which has been reviewed by Lieber et al., is another major pathway for repairing DSBs. In common with HR, this pathway is important in the repair of agents that result in DSBs such as IR, bleomycin, topoisomerase II poisons, and anthracyclines. The DNA-dependent protein kinase (DNA-PK) consists of the catalytic subunit (DNA-PKcs) and the regulatory subunit (the Ku70/Ku80 heterodimer). The DNA-PKcs subunit is a serine/threonine kinase that belongs to the phosphatidylinositol-3 kinase family. The Ku80/Ku70 heterodimer (Ku) exhibits sequence-independent affinity for double-stranded termini and, on binding to DNA, recruits and activates the DNA-PKcs catalytic subunit.
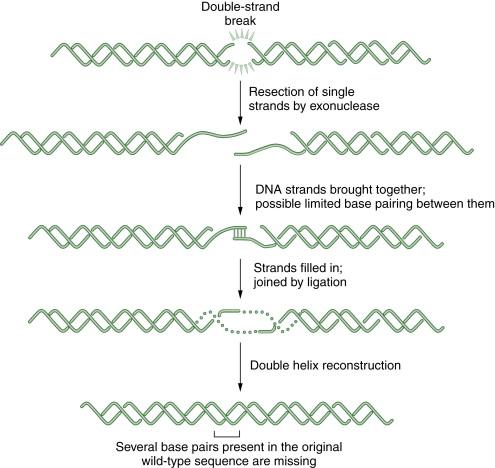
Additional proteins are required for the completion of NHEJ, including the Artemis protein and DNA ligase IV. Importantly, NHEJ is an error-prone repair pathway. Because the process does not use a complementary template, the fusion of the blunt-ended DNA duplexes may result in deletion or insertion of base pairs. A schematic representation of NHEJ is shown in Figure 4-5 . NHEJ has a normal function in immune cells to generate diversity at the immunoglobulin and T-cell receptor gene loci.
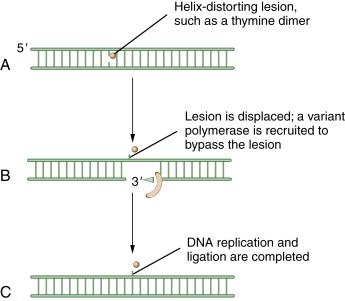
The process of TLS is another mechanism for dealing with thymine dimers and bases with bulky chemical adducts. At a DNA replication fork, DNA adducts may cause a replicative polymerase such as DNA polymerase delta to stall. Cells have, therefore, developed sophisticated mechanisms for switching off the replicative polymerase and switching on alternative polymerases (i.e., a polymerase such as pol eta, which will replicate past certain DNA lesions with high fidelity). Interestingly, human cells have at least 15 DNA polymerases, although the situations and mechanisms of their deployment are largely unknown. Cancer may have a heightened dependence on one of the error-prone TLS polymerases, such as polymerases β or kappa, accounting for high rates of mutagenesis. A schematic representation of TLS is shown in Figure 4-6 .
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here