Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The discovery of cancer susceptibility regions across the genome provides opportunities to understand defining events in tumor development, specifically identifying cellular pathways that contribute to the complex development of cancer.
Regions of the genome harboring susceptibility alleles and genes can be identified in families or unrelated populations by using association studies, next-generation sequencing, and linkage studies.
Technologic advances in sequence analysis in concert with comprehensive annotation of genetic variation across the human genome continue to accelerate the pace of discovery and characterization of cancer susceptibility alleles. The conclusive identification of a gene or a regulatory region contributes to an understanding of defining events in tumor development.
The spectrum of cancer susceptibility alleles includes mutations in genes that are highly penetrant, which indicates that individuals born with a mutant allele have a high probability of developing cancer; moderately penetrant mutations, which confer an increase in probability; and common variants, which impart a small risk for cancer.
Both association studies and family-based studies require accurate collection of clinical data by clinicians, and both represent an important pillar for precision medicine and precision prevention. An improved understanding of molecular aspects of cancer and specifically the use of biomarkers such as susceptibility alleles can inform clinical and public health decisions.
For generations, investigators have pursued the heritable contribution to cancer. Seminal studies in families in which members affected with breast cancer, colorectal cancer, melanoma, or a constellation of cancers (e.g., Li-Fraumeni syndrome) have provided evidence for rare mutations with strong effects. Generations of family-based and twin studies have repeatedly shown an excess of familial cancer aggregation for nearly all types of cancers, although the estimates vary greatly across cancer types. These observations suggested that it would be possible to map cancer genes and thus estimate the genetic contribution to each molecular type of cancer, even in unrelated populations. Until the mid-2000s, progress had been slow. However, the pace at which new genetic regions harboring cancer susceptibility alleles have been identified has accelerated substantially because of three converging factors. First, a high-quality draft sequence of the human genome was produced. Second, its subsequent annotation resulted in the appreciation of a wide spectrum of variation across the genome. Third, the development of technical platforms that enable interrogation of genetic variation across the genome has changed the discovery speed of cancer susceptibility alleles. In the last decade the scope of studies changed dramatically, expanding from family-based studies to larger population-based studies of unrelated individuals. These studies have been fueled by the precipitous drop in price for interrogating large numbers of informative single-nucleotide polymorphisms (SNPs), the most common type of variant in the genome, or massive parallel sequence analysis of entire or partial genomes, particularly the coding regions of the genome (known as the exome ). To keep pace with new streams of large data sets, investigators have forged new collaborations and developed computational tools for analyzing ever-enlarging data sets in search of new cancer susceptibility alleles. The ability to discover and validate cancer susceptibility alleles is highly dependent on sharing of data through accessible databases enabling bona fide and approved researchers to carry out additional studies.
Cancer susceptibility alleles have been discovered through a spectrum of approaches, yielding a range of inherited genetic variants from rare mutations with strong effects (i.e., highly penetrant) to common genetic polymorphisms, each of which confers a small risk for cancer. By definition, susceptibility alleles increase an individual's risk of developing cancer either within families or across populations, but there are instances of pleiotropy in which a particular variant may confer susceptibility to one type of cancer yet protect against another type of cancer. It is notable that not all susceptibility alleles for a given gene have equal estimated effects. Consequently, the observed spectrum of established susceptibility alleles reflects an inverse relationship between the effect size and the frequency of the genetic variation ( Fig. 21.1 ). So far, approximately 125 hereditary cancer predisposition genes (CPGs) have been identified, and their presence is most evident in studies with familial clustering of one or more cancers. Moreover, the majority of the hereditary predisposition genes are known somatic mutational drivers of cancer, highlighting an important principle first outlined by Knudson in 1971. The first set of CPGs was discovered in family studies using linkage analysis, but now next-generation sequencing (NGS) analysis has accelerated the discovery of new genes and specific mutations in known CPGs. Susceptibility alleles that have an appreciable frequency in the general population, and smaller effect sizes, can be discovered through association studies in which the genomes of a set of affected cases are compared with those of unaffected cancer-free or population-based controls.
Genetic mapping of cancer susceptibility genes has shown that many signals map to nongenic regions, important in the regulation of genes or pathways of interacting genes. Although the direct public health impact associated with conclusively establishing a specific cancer susceptibility allele may not be immediately apparent, its contribution to understanding tumor development and metastasis is invaluable, expanding possible pathways and putative targets for intervention downstream. Moreover, the possible clinical value of known susceptibility alleles will continue to increase as more comprehensive maps of susceptibility alleles emerge for specific cancers. In this regard, sets of variants tested together show great promise for risk stratification, important for both the individual and at the population level. Roughly 10% to 15% of cancer susceptibility alleles are shared among cancer types, and in some circumstances there are distinct differences by subtype (e.g., specific alleles for estrogen-negative breast cancer). To define the genetic architecture (see Fig. 21.1 ), namely the constellation of susceptibility alleles that contributes to a specific cancer, continued efforts are required to define comprehensive sets of variants, which in turn should emerge as vital tools in both public health and individual (known as precision medicine ) assessments of cancer risk.
The annotation of sequence variation in the genome has provided important clues toward elucidation of the genetic history of distinct populations, the possible impact of environmental or pathogenic assault on the genome, and the heterogeneous distribution of human cancers. The differences in allele frequency spectrum, as well as types of genetic variation, from SNPs to large copy number variants, have become indispensable tools for geneticists in identifying cancer susceptibility alleles ( Fig. 21.2 ).
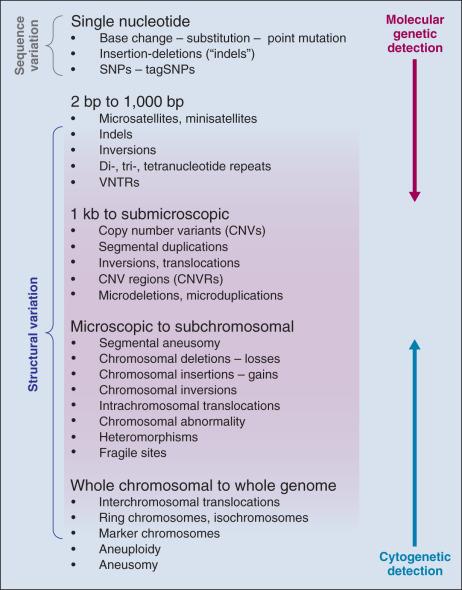
The search for common alleles is predicated on conclusively finding “markers” that highlight a region of the genome where a disease susceptibility allele resides; subsequent mapping and laboratory studies are required to understand the link between the susceptibility allele and its biologic association with disease. Sets of markers to test are drawn from dense maps of human genetic variation that are publicly available. Although there has been great value in embracing this indirect approach ( Fig. 21.3 ), it comes at a price—namely, many additional steps are needed in order to sort through the correlated variants and then conduct the functional studies that are necessary to illuminate the underpinnings of the susceptibility allele.
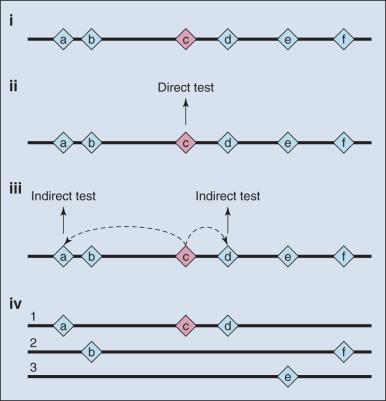
Until whole human genome sequence became available, the field of genetics created maps of variant coordinates based on incomplete assembly of DNA segments spanning the human genome. Sets of markers can be thought of as molecular street signs, which allowed scientists to knowingly navigate their way up or down a region of a chromosome. Early on, “genetic maps” provided a stable reference for mapping highly penetrant mutations, primarily in families. These were based on empiric evidence of recombination hot spots. The long-standing value of functional elements, herein recombination frequencies, served adequately for the mapping of disease and traits until genome sequencing emerged as a viable tool. The emergence of a physical map of the human genome that establishes the true order of most DNA segments (currently tractable for more than 95% of the genome) has accelerated the mapping of traits and diseases, as it produced absolute coordinates, or street signs for variants within the genome—that is, the nucleotide location of a given marker or gene in millions of base pairs from the end or terminus of the chromosome is generally known.
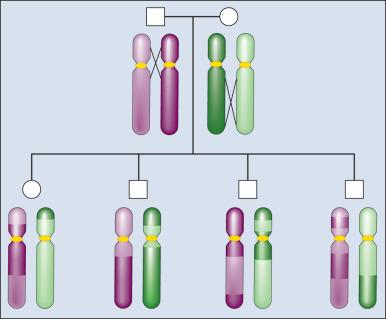
The principle of meiotic recombination is critical to understanding the relationship between genetic loci, here defined as variants that map to unique coordinates. The correlation between genetic markers is fundamental to both association and linkage analysis. In meiosis, the cell division leading to gamete formation pairs homologous chromosomes. Each chromosome consists of two identical strands (chromatids), with each chromosome pairing composed of four strands. Homologous chromosomes separate from each other during the process of meiosis except at one or two zones of contact in a process that leads to genetic recombination ( Fig. 21.4 ). Mendel's second law, independent assortment, states that alleles of genes at unlinked loci, such those on different chromosomes or at the ends of a chromosome, segregate or assort independently. Deviations from independent assortment occur when genes are located close to one another, in which case alleles assort together more than 50% of the time. In this scenario, the associated loci are “linked.” Distributed throughout the genome are recombination hot spots, which “divide” the genome during meiotic formation of egg and sperm. These can vary by population genetic history, providing an opportunity to compare groups and use the differences to pinpoint possible susceptibility alleles, especially if there are substantive differences between populations with respect to cancer incidence.
The spectrum of human genetic variation varies by the frequency of polymorphisms, which is often substantial among populations. The most common sequence variation is the substitution of a single base, known as a single-nucleotide polymorphism, which, by definition, must be observed in at least 1% in one or more populations. The minor allele frequency (MAF) refers to the lower allele frequency, often varying by population ancestry. A substantially larger fraction of genetic variation exists for single base substitutions below 1%—and many of these are population private, reflecting population genetics history. The majority of SNPs with an MAF greater than 10% are common to most human populations, but the actual frequencies can vary. Reported SNPs are cataloged in db-SNP ( http://www.ncbi.nlm.nih.gov/snp ), which is an important reference that is useful for interpreting variants identified through DNA sequencing.
A small subset of SNPs are located in exons, of which a fraction change the predicted amino acid. SNPs that can alter the coding sequence are known as nonsynonymous SNPs, whereas those that are silent are termed synonymous. Although there has been great interest in coding SNPs, partly because they appear to be more interpretable, very few of the known associations between a disease and a common SNP marker (MAF > 10%) involve coding SNPs. On the other hand, rare highly penetrant mutations mainly map to coding changes or preterminal stop codons. Many of the reported disease mutations, known as cancer predisposition genes, are cataloged in public databases such as Online Mendelian Inheritance in Man ( https://www.omim.org/ ) or Catalogue of Somatic Mutations in Cancer (COSMIC; http://cancer.sanger.ac.uk/cosmic ).
SNP frequencies become fixed in populations over multiple generations and are generally not inherited independent of the adjacent variants. Recombination hot spots can separate sets of highly correlated variants, resulting in haplotype blocks ( Fig. 21.5 ). These segments of a chromosome, usually quite small, are transmitted as a unit from one generation to the next. The correlation between SNPs is an estimate of linkage disequilibrium (LD), which is classically defined as the nonrandom association of alleles at different loci. Individual SNPs that always track together are said to be in strong LD. This correlation can be eroded over time by recombination (exchange of genetic material) during meiosis, and SNPs can be defined as being in weak LD, signaling a correlation that is not necessarily strong. Measurement of LD is with either D′ or r 2 statistics.
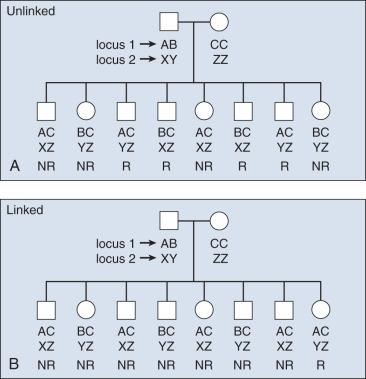
The concept of LD is important because it enables investigators to evaluate sets of SNPs and determine proxies for other, untested SNPs in a region; this is useful for indirect mapping. If a group of SNPs is in strong LD (e.g., they are inherited together), one can test for the alleles of just one reference SNP and immediately have information regarding alleles segregating in a given individual for all adjacent, correlated SNPs. By extension, estimates of LD are useful to construct haplotypes in unrelated subjects. With new reference data sets (e.g., 1000 Genomes Project), it is possible to reliably determine or “impute” untested variants using the backbone of stable data sets. Computational efficiencies enable estimation of the correlation between sets of markers and the construction of haplotypes. Still, the most reliable approach is to resolve the phase of haplotypes in multigeneration pedigrees, in which haplotypes can be traced; alternatively, one can infer the relationship of alleles in unrelated subjects with computational tools. Phase refers to the parental (and grandparental) chromosome of origin for a set of alleles. This information can also be useful for determining where in a family a disease allele originates.
The annotation of the human genome has revealed a wide spectrum of structural variations, which may be either cytologically visible or detected with either microarray chips or actual sequence analysis (see Fig. 21.2 ). Historically, short tandem repeats (STRs) are a class of polymorphisms in which there is a reiteration of a small number of base pairs, such as CACACA. Polymerase chain reaction (PCR) primers are used to define the distinct physical location of one STR from the remaining 50,000 in the genome and to ascertain the number of repeats for both chromosomes for any given STR. Also known as microsatellites, they have been used for linkage analysis and forensic investigation. Structural variants of all sizes can include deletions, insertions, and duplications collectively known as copy number variations (CNVs). In addition, there are infrequent inversions and translocations of pieces of DNA that vary in size. Some of these are quite common; chromosome 17 harbors an inversion of 3.5 million base pairs in approximately 20% of the European population. CNVs have been shown to influence gene dosage and therefore can contribute to risk for cancer, as demonstrated for a chromosome 1 CNV and risk for childhood neuroblastoma. There have been formidable technical challenges in calling CNVs, but with new resources and new sequencing technologies termed next-generation sequencing, it is anticipated that accuracy will continue to improve, which in turn should improve the capacity to associate CNVs with disease outcomes.
Many epidemiologic studies have indicated the presence of a familial contribution, such as the observation that family history of a specific cancer within first-degree relatives is associated with a doubling of risk or greater among relatives, particularly in twin registries. In the case of prostate cancer, for instance, studies of selected hospital-based patient populations, population-based case-control studies, and cohort studies all have demonstrated that a family history of disease is correlated with an increase in an individual's risk. If the affected family members are first-degree relatives (i.e., brothers, fathers, sons), the risk increases from 1.7-fold to 3.7-fold. Younger ages at diagnosis and multiple affected relatives with the disease tend to be associated with even higher relative risk (RR). For example, men with three or more first-degree relatives with prostate cancer have an almost 11-fold increased risk of the disease compared with men who have no known family history of the disease. For this reason, families ascertained for linkage analysis studies tend to be large, have multiple affected individuals, and feature people who were diagnosed with the disease at a comparatively young age.
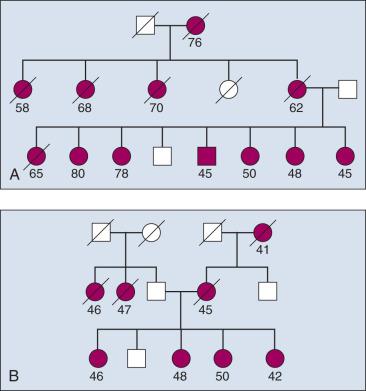
Familial aggregation describes the occurrence of multiple cases of cancer within a family ( Fig. 21.6 ). Clustering of familial cases can also be due to shared environment, shared alleles of particular genes, or simply chance if the tumor is common in the population. In mapping cancer susceptibility genes for many cancers, particularly for breast cancer and colon cancer, the most promising pedigrees for hereditary cancer are families with three or more first-degree relatives with a given cancer, three successive generations with cancer, or at least two siblings with the same cancer detected at a relatively young age. For common cancers, details regarding tumor stage and grade at diagnosis, histopathology, and response to treatment are key to linkage analysis study design because individuals who share similar disease features likely share common susceptibility alleles. For instance, if a subset of affected individuals in the family in Fig. 21.7 all had tumors of similar stage and grade, the data from this homogenous subset of individuals could be considered in isolation from the rest of the affected cases thus reducing heterogeneity and increasing power. For common diseases there are many common susceptibility alleles that account for a substantial fraction of the underlying genetic architecture of cancer susceptibility, and this approach generally does not work. However, it has proven useful in discovery of highly penetrant alleles for many cancers (e.g., breast and colon cancers).
To identify highly penetrant mutations, success directly correlates with the identification and collection of high-risk or hereditary families. To achieve the numbers needed to improve the power to detect a disease allele, whether using SNP arrays or NGS, large consortia are often formed, providing an opportunity to increase power through addition of more families and the chance to define the phenotype—namely, the required clinical features and family history. Large consortium studies provide an opportunity to conduct segregation analysis, which determines the most likely genetic model that could account for the disease (e.g., dominant, codominant, recessive, or sex-linked). Additional informative analyses include an estimate of the frequency and penetrance of the disease allele(s) in the general population, age-dependent penetrance, and potential number of loci contributing to the disease.
High-risk or hereditary families must be ascertained through use of appropriate guidelines for working with human subjects to collect biospecimens, such as germline DNA from blood or buccal materials, somatic or tumor tissue for DNA or RNA analyses, and other body fluids for determination of biomarkers that could be useful in subsequent early detection in high-risk settings. Identification of cancer families and collection of critical medical information including family history, medical record data, and DNA samples are generally regulated by institutional review boards (IRBs) and require informed consent. Families must be identified and approached in a way that is neither intrusive nor coercive. Although genetic epidemiologists have successfully turned to new approaches, such as social media, to ascertain families, most families are made aware of or referred to studies by their personal physicians.
To carry out a successful family-based genetic study, medical record data must be carefully and systematically extracted into well-protected databases. Family history data must also be obtained redundantly from multiple family members, with care taken to resolve discrepancies, including nonpaternity events. Consent to contact other family members regarding the study is needed, as is permission to obtain medical records and to recontact study participants years after initial data collection. The protection of individual privacy is paramount, and personal identifiers such as names and complete addresses are rigorously protected, even from most of the scientists participating in the study.
DNA samples from appropriate family members can be screened by using either a set of highly polymorphic markers that span the genome at a sufficiently high density or NGS analysis of the whole genome or the exome.
Theoretically, a given set of affected individuals within a family would all carry precisely the same underlying mutation in the relevant susceptibility gene—that is, each member would have inherited not just a mutated copy of the same gene, but the same deleterious variants in that gene. Because there are distinct mutations within a gene, each of which can confer high penetrance, the approach is predicated on finding a gene and not the specific mutation within a gene. For example, there are many mutations and variants of unknown significance across the BRCA1 gene that can confer an increased risk for breast and/or ovarian cancer, with measurable differences in penetrance now confirmed by prospective studies. This latter point suggests that there are differential effects of disturbances of key biologic pathways. Moreover, recent genome-wide association studies (GWASs) have begun to identify secondary, genetic modifiers that further modulate the penetrance of BRCA1 mutations.
Fig. 21.7 demonstrates two types of seemingly useful families for linkage mapping studies. Both include a significant number of affected members. The first family has a large number of affected individuals (see Fig. 21.5 ). However, some individuals were affected very early in life, whereas others were diagnosed at later ages. It is likely that some individuals have the disease because they inherited mutated copies of a particular gene, whereas others have the disease for sporadic reasons unrelated to the disease allele segregating in the family. Age at onset provides guidance as to which individuals are more likely to have hereditary versus sporadic forms of the disease; but this is not absolute, and in the case of a disease with age-dependent penetrance, some people will be affected late in life, even though they carry a mutant allele, and others will be affected early in life for sporadic reasons. The second family shown in Fig. 21.5 appears to be more informative for linkage mapping studies because there are several affected individuals in the family and all were affected at a relatively early age. However, the presence of disease segregating on both sides of the family should be noted. The affected individuals in the youngest generation could have cancer because they inherited mutant alleles from one or both sides of their family, and one or multiple genes could be involved. This scenario happens frequently in studies of common cancers such as those of the colon, breast, and prostate. Thus the family is of limited usefulness for mapping studies.
Traditional approaches have been successful in identifying highly penetrant mutations in multiply affected families for both common and uncommon cancers. A combination of linkage and candidate gene analyses revealed mutations in CDKN2A or CDK4 in roughly 50% of familial melanomas, although there appears to be heterogeneity in exposure to a strong carcinogen for melanoma, ultraviolet sunrays. Newer approaches involving analysis of smaller family structure together with functional laboratory data have identified rare, important mutations in melanoma. For a rare familial cancer, chordoma, a gene duplication of the T (brachyury) gene confers susceptibility. Using NGS, investigators are revisiting families whose disease was not understood when traditional methods were used, and are now searching for sets of susceptibility alleles that can explain an oligogenic risk model—that is, a set of variants with moderate risk that are neither sufficient nor necessary for development of a cancer (e.g., MITF is a moderate-risk gene for melanoma).
The early-onset breast cancer susceptibility genes BRCA1 and BRCA2 were among the first to be mapped because large and well-characterized families had been meticulously ascertained. In addition, deleterious alleles were highly penetrant, often at an early age, leaving little ambiguity as to who in a family should be counted as a “case.” The presence of ovarian cancer in some families and not others and the presence of breast cancer in some male carriers allowed for creation of data sets enriched for the BRCA1 and BRCA2 genes, respectively. In turn, the initial identification of the BRCA1 gene and subsequent removal of BRCA1 -linked families from remaining data sets provided further useful enrichment for BRCA2 -linked families. For the breast cancer susceptibility genes BRCA1 and BRCA2, founder mutations have been identified in distinct populations across the globe, a phenomenon most notably observed in Jewish Ashkenazi families. The three common founder mutations in this population, BRCA1-185delAG, BRCA1-5382insC, and BRCA2-6174delT, have a combined population prevalence of 2% to 2.5%. With these observations in mind, investigators have frequently sought families for genetic mapping studies from regions of the world where marriage between related individuals is not discouraged and where geographic barriers have restricted gene flow.
Locus heterogeneity, the presence of deleterious alleles associated with many genes in a population, can be reduced by studying families from isolated or inbred populations. Fewer disease alleles are predicted to segregate with a particular phenotype in a population derived from a limited number of founders. Studies of colon cancer in Finland and breast cancer in Iceland as well as Ashkenazi Jewish populations illustrate this point. In Finland, two variants in the DNA mismatch repair gene MLH1— mutations 1 and 2—account for 51% of all Finnish families with verified or putative cases of hereditary nonpolyposis colorectal cancer. Nineteen mutation 1 and six mutation 2 families were further investigated by haplotype analysis using 15 microsatellite markers surrounding the MLH1 locus. The presence of two distinct large conserved disease haplotypes, one in mutation 1 and the other in mutation 2 families, indicated that these families are likely to descend from two distinct common ancestors born in the 16th and 18th centuries, respectively.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here