Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
In December 2019, a cluster of patients with pneumonia of unknown etiology were identified in Wuhan, a city of 11 million residents in South Central China. RNA extracted from bronchoalveolar fluid provided the complete genome of a novel betacoronavirus named SARS-CoV-2 (for severe acute respiratory syndrome coronavirus-2). The disease caused by this virus is called COVID-19 (for COronaVirus Infectious Disease).
SARS-CoV-2 is classified within the Coronaviridae, a diverse family of viruses with (+) strand RNA genomes ( Table 335-1 ). Coronaviruses, which infect not only humans but also other mammalian and avian species, may cause respiratory or enteric diseases. SARS-CoV-2 is the seventh coronavirus known to infect humans. Four of these human coronaviruses (HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1) cause mainly mild upper respiratory tract infections ( Chapter 334 ). By comparison, severe acute respiratory syndrome coronavirus (SARS-CoV), which was identified in 2002, precipitated an epidemic that infected about 8000 humans before its circulation ceased. Middle East Respiratory Syndrome coronavirus (MERS-CoV) was first identified in 2012 and continues to infect humans, although chains of infection are very short, and only about 2500 confirmed cases have been reported.
| FAMILY: CORONAVIRIDAE | |
| Genus: Alphacoronavirus | Viruses: Human coronavirus 229E (HCoV-229E) Human coronavirus NL63 (HCoV-NL63) |
| Genus: Betacoronavirus | Viruses: Human coronavirus HKU1 (HCoV-HKU1) Severe acute respiratory syndrome coronavirus (SARS-CoV) Middle East respiratory syndrome coronavirus (MERS-CoV) Severe actute respiratory syndrome coronavirus 2 (SARS-CoV-2) |
∗ The Coronaviridae includes five genera, of which two, the Alphacoronavirus and Betacoronavirus , include the six known human coronaviruses.
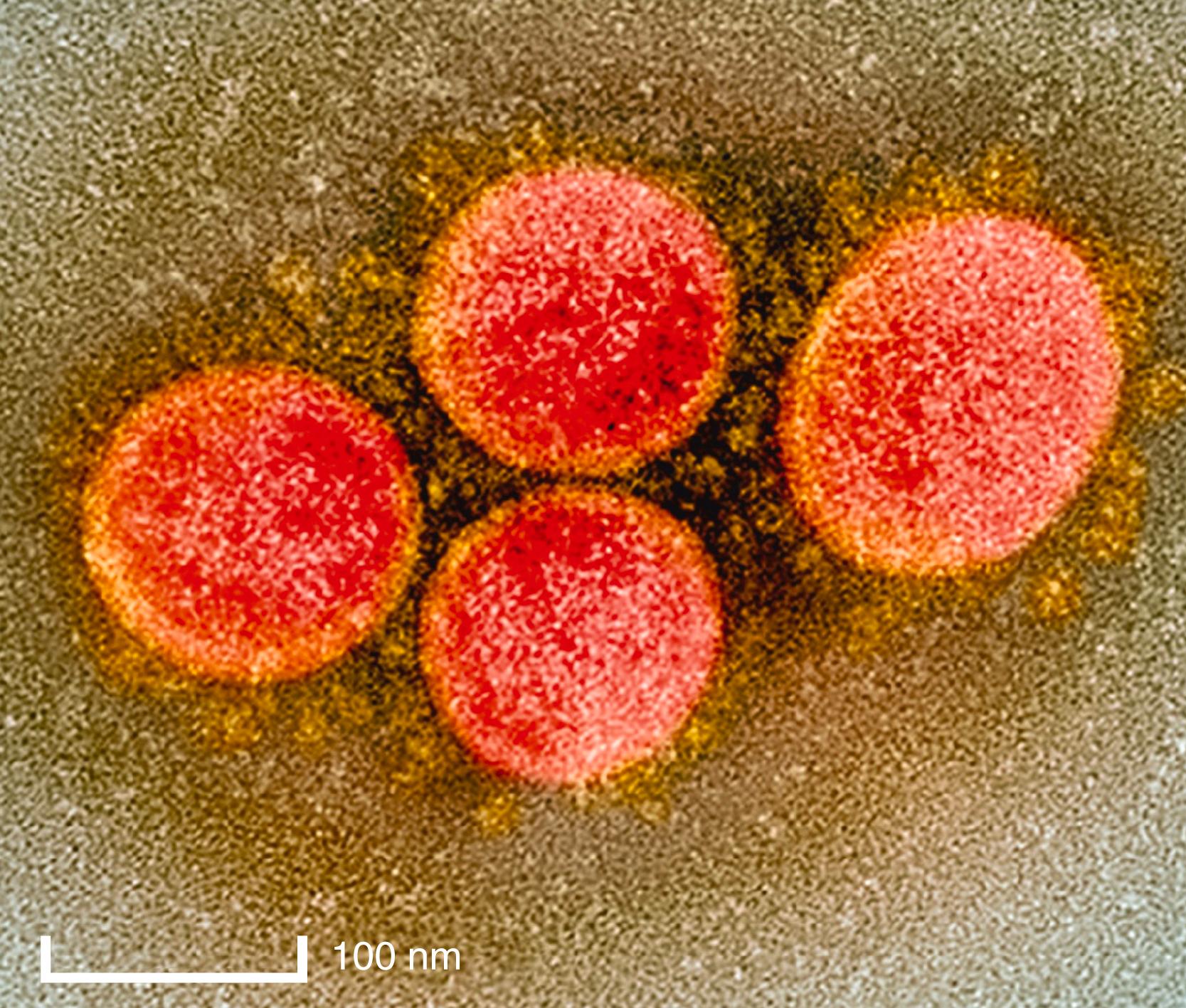
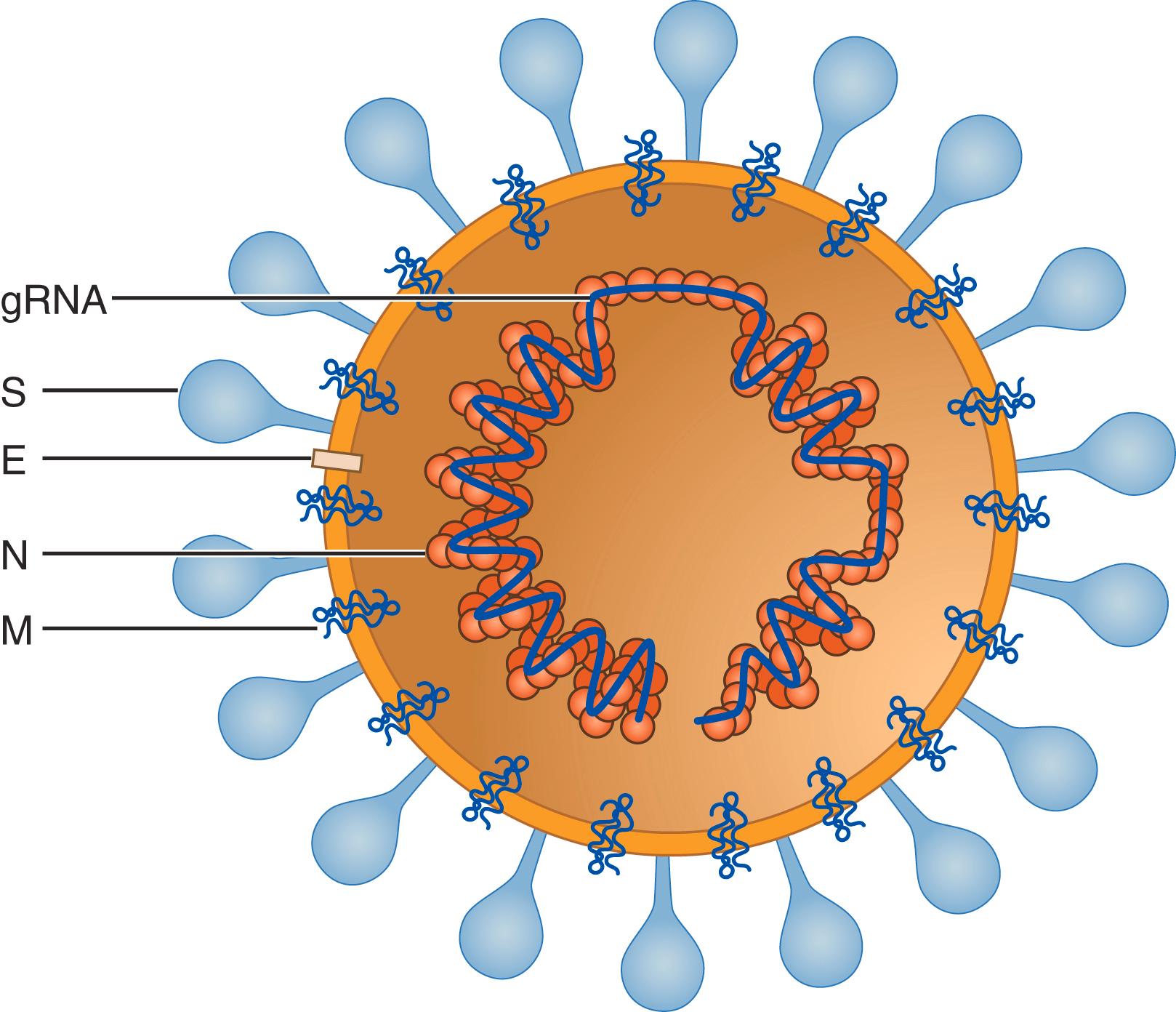
Coronaviruses, which have the appearance of a solar corona on electron microscopy ( Fig. 335-1 ), are enveloped, roughly spherical, and pleomorphic, with particle sizes ranging from approximately 118 to 136 nm in diameter. Embedded in the viral membrane are the spike (S), membrane (M), and envelope proteins ( Fig. 335-2 ). The transmembrane S protein forms trimers that bind to host cell receptors and induce fusion of viral and cell membranes. The M protein monomer, which provides shape to the viral particles, is embedded in the viral membrane by three transmembrane domains. The E protein is a small (8- to 12 kDa) protein that is essential for infectivity, but its function is enigmatic. The nucleocapsid (N) protein, which is the most abundant protein in the infected cell, is required for replication of the genome, coats the entire viral RNA genome, and interacts with the M protein.
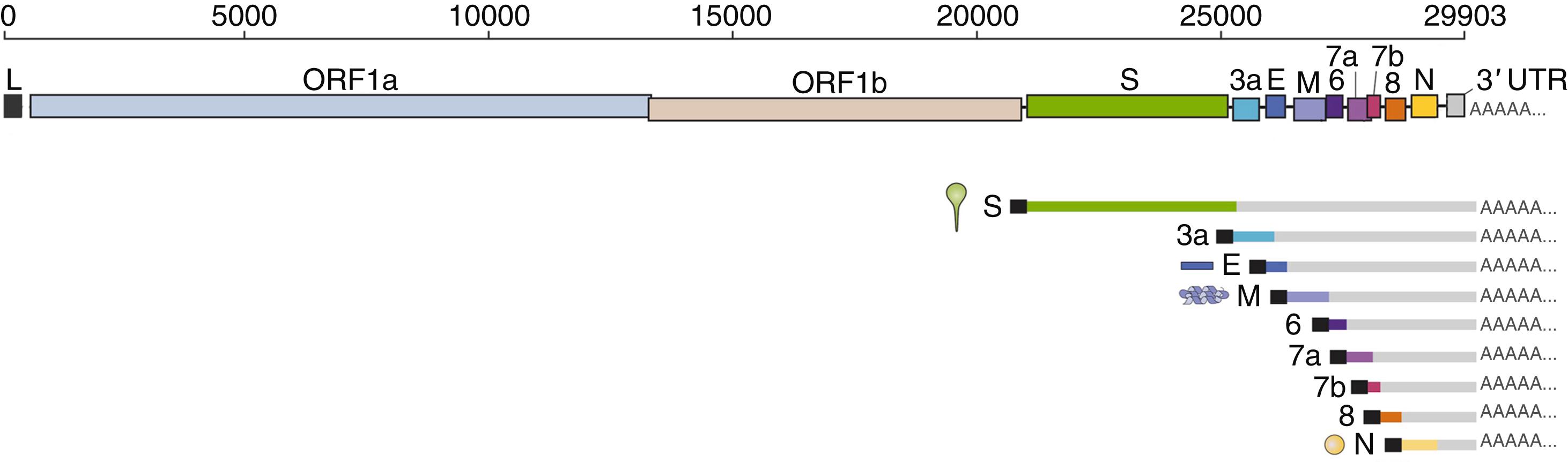
Coronavirus genomes are the longest among RNA viruses, and the SARS-CoV-2 genome is nearly 30,000 bases in length ( Fig. 335-3 ). The virion RNA is capped at the 5ʹ-end and polyadenylated at the 3ʹ-end and harbors 5ʹ- and 3ʹ-untranslated regions. The left two-thirds portion of the viral RNA encodes two overlapping open reading frames that specify proteins involved in mRNA synthesis and genome replication; no other polypeptides are translated from the viral genome RNA. The remainder of the genome, which encodes structural and accessory proteins, is expressed by means of a nested set of subgenomic RNAs.
The closest genome to SARS-CoV-2 is a virus called RaTG13, which was obtained from a Rhinolophus affinis bat in 2013 in Yunnan province, China. The genome of this bat virus is overall 96.2% identical to that of SARS-CoV-2, a divergence that represents about 20 years of sequence evolution. Consequently, RaTG13 cannot be a direct progenitor of SARS-CoV-2 but is clearly in its evolutionary past.
Various SARS-CoV-2–like viruses have been isolated from bats in China, Thailand, Cambodia, and Japan, but none have a spike protein that can bind angiotensin-converting enzyme 2 (ACE2) and allow entry into human cells. In limestone caves in Northern Laos, however, sampling of bats has revealed the presence of three Sarbecovirus genomes (called BANAL-52, BANAL-103, and BANAL-236) with high sequence similarity to SARS-CoV-2 and RaTG13. Of the 17 amino acids that interact with the receptor-binding domain of ACE2, 16 are conserved between SARS-CoV-2 and BANAL-52 or BANAL-103, and 15 of 17 are conserved with BANAL-236. In contrast, only 11 out of 17 receptor-binding domain amino acids are conserved among SARS-CoV-2 and RaTG13. Recombination analysis demonstrates that the SARS-CoV-2 genome is a mosaic of at least five different genomes, including BANAL-52, BANAL-103, and BANAL-236 and the previously recognized RmYN02, RpYN06, and RaTG13, the latter all discovered in China. The implication of these observations is that SARS-CoV-2 likely arose from recombination among viruses circulating in different species of Rhinolophus bats in the limestone caves of South China and Southeast Asia.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here