Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Dedicated to the loving memory of Eta Najfeld, MD, a Holocaust survivor and an amazing mother .
Over the past 66 years the cytogenetic analysis of hematologic malignancies has been an area of prolific growth. Chromosome studies and karyotype analysis provide information of both biologic and clinical significance. Refinements in cell culture methods and the application of chromosome banding techniques have advanced our understanding of disease-specific abnormalities, and molecular cytogenetic and cytogenomic methods now have made possible the identification of genes involved at translocation breakpoints in specific chromosomal rearrangements. These advances in molecular cytogenetic methods permit mapping of structural rearrangements within a single gene and have fundamentally contributed to our knowledge of the biology of many hematologic malignancies. This evolution in our understanding of cancer genetics has resulted in a distinct terminology ( Table 57.1 ). Application of conventional and molecular cytogenetic methods has identified over 100 fusion genes involving over 500 different genes and approximately 1000 recurrent balanced translocations in human cancers. Relevance of these methods has played a pivotal role in the diagnosis, treatment, and prognostications of the hematologic malignancies. This chapter discusses specific cytogenetic events and delineates molecular phenotypes that are important to understand the molecular pathogenesis of hematologic malignancies and provides several genetic testing algorithms. The remarkable hypothesis put forward by Boveri at the turn of the 20th century—namely, that an abnormal chromosome pattern is intimately associated with the malignant phenotype of a tumor cell—has proven correct. Knowledge of the molecular cytogenetic genotype of hematologic malignancies has led to innovative and targeted treatments. The first example of such gene-targeted therapy has already been successfully applied to chronic myeloid leukemia (CML) ( Chapter 69 ).
| Aneuploidy | Abnormal chromosome number, either gain or loss. |
| Array CGH | A higher-level CGH technology that provides gene copy information. |
| Balanced translocation | Exchange of chromosomal material that creates no extra or missing DNA. |
| Banding |
|
| Breakpoint | Specific site on a chromosome containing a break in the DNA that is involved in chromosomal structural rearrangement, such as translocation or deletion. |
| Centromere |
|
| Centromere enumeration probe (CEP) | Highly repetitive α (or β) satellite DNA, located in the heterochromatin of the centromeric area of chromosomes. CEP targets repetitive α (or β) sequences produces bright compact fluorescent signals. |
| Comparative genomic hybridization (CGH) | Molecular cytogenomic technique that provides a copy-number karyotype at the chromosome and band level. |
| Chromosomal rearrangement | Aberration in which chromosomes are broken and rejoined. |
| Clonal abnormality | In cytogenetic analysis, two cells showing the same additional or structural abnormality or three cells with loss of the same chromosome. In FISH analysis, any abnormality present after the probe has been validated and normal reference range established, above the normal reference range. |
| Chromothripsis | A catastrophic DNA damage occurring during a single mitotic division the application of molecular biology to determine genomic copy number. |
| Deletion | Segment of chromosome that is missing (terminal) or segment of chromosome missing between two breakpoints (interstitial). |
| DNA sequence | Order of nucleotides in a DNA segment, usually displayed from the 5′-triphosphate (5′ end) to the 3′-hydroxyl (3′ end) nucleotides. |
| Driver mutation | This mutation affects the biology of cell. |
| Enhancer | DNA sequence that increases the rate of transcription. |
| Exon | Portion of gene that encodes protein. |
| Fiber FISH | Application of FISH technology to extended DNA or free DNA fibers. |
| FISH | Fluorescence in situ hybridization, a method for detection of the number and location of DNA sequences (genes) in tissue section or cell population. |
| Fluorochrome |
|
| Gene construct | Recombinant DNA containing a gene of interest surrounded by sequences engineered to promote a measure of its expression. |
| Gene map | Order of genes within a chromosome or entire genome. |
| Genotype | Genetic constitution, usually with reference to particular alleles at a locus. |
| Haploid | Half of a normal complement (i.e., 23 chromosomes). |
| Haploinsufficiency | Deletion or inactivation of one allele producing disease caused by inadequate activity of the remaining allele. |
| Hybrid gene | Fusion of two different genes as a result of a structural chromosomal rearrangement that functions as one transcriptional unit. |
| Hybridization | Method for rejoining (reannealing) complementary DNA or RNA strands. |
| Hyperdiploid | Additional chromosomes (e.g., 47 or 48 chromosomes). |
| Hypermetaphase FISH | Application of FISH to accumulated large number of metaphase cells. |
| Hypodiploid | Loss of chromosomes (e.g., 45 or 44 chromosomes). |
| I-FISH | Interphase fluorescence in situ hybridization, application of FISH to nondividing (resting) cells. |
| Interphase | Stage of mitosis in which the cell is not dividing. |
| Inversion |
Pericentric inversion refers to breaks occurring on the opposite side of the centromere. |
| Isochromosome | Structural chromosomal rearrangement that consists of doubling of one of the two chromosome arms (connected by the centromere) and loss of the other arm. |
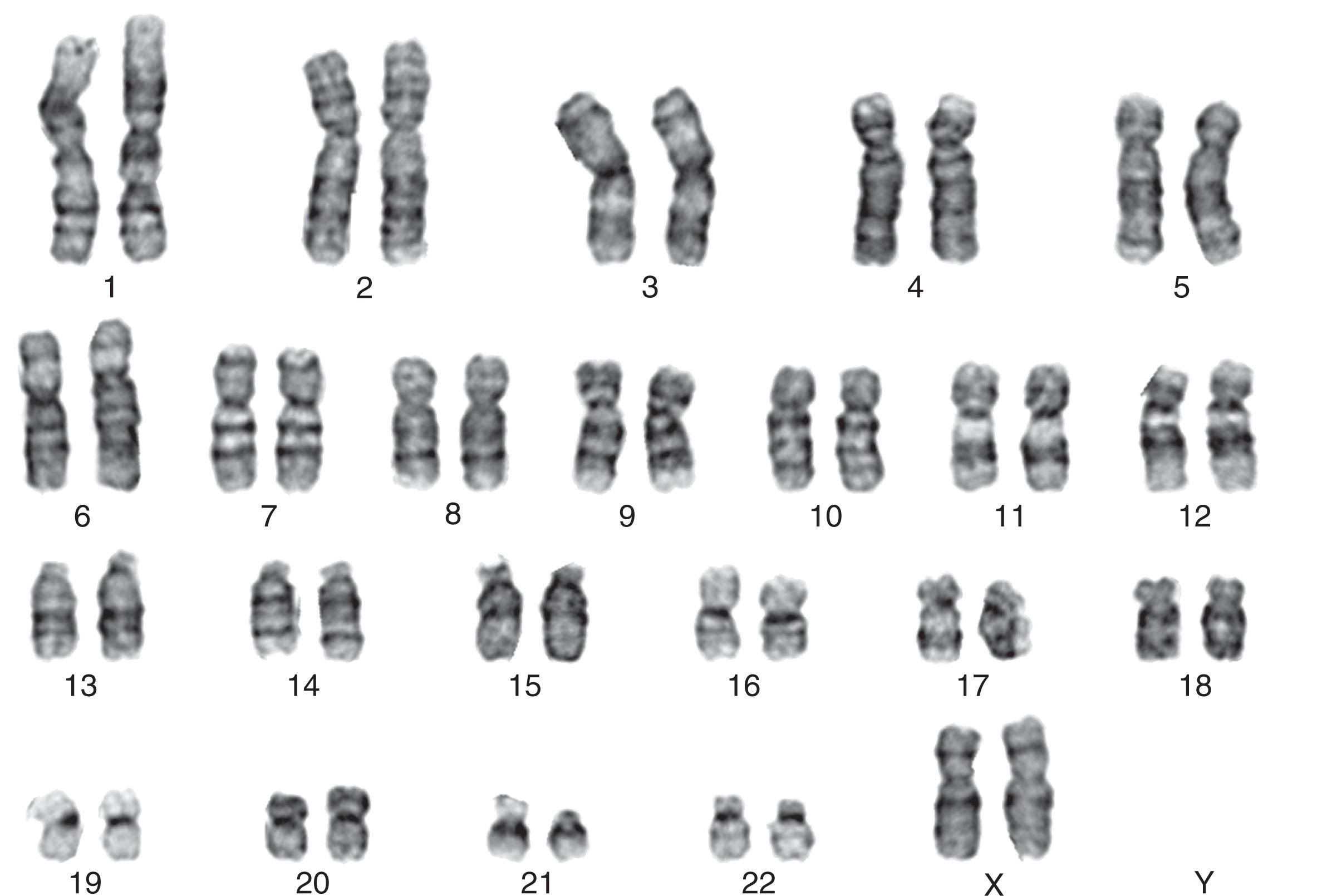
| Karyotype | Arrangement of metaphase chromosomes from a particular cell according to size and banding so that the largest chromosome is placed first and the smallest one last (see Fig. 56.2). |
| kb (kilobase) | Unit of DNA/RNA length = 1000 base pairs of DNA. |
| Kataegis | Kataegis is a recently discovered phenomenon in which multiple mutations cluster in a few hotspots in a genome. This region is often colocalized with regions of somatic genome rearrangements. |
| Locus | Unique location of a gene on a chromosome. |
| Locus (sequence)-specific probe (LSI) |
|
| Marker chromosome |
|
| M-FISH | Multicolor FISH karyotyping, which allows identification of 24 different human chromosomes (22 autosomes, and the X and Y chromosomes) (see text for details). |
| NGS | Next generation sequencing. |
| Nonsilent mutations | Mutations that alter the protein sequences. |
| Oncogene | Locus that is activated in association with tumor growth. One abnormal allele is sufficient to cause tumor formation or cancer. |
| SNP | Single nucleotide polymorphism. |
| Passenger mutation | Is usually a subclone and does not affect the biology of cell. |
| PCR | Polymerase chain reaction, by which individual gene segments are amplified through sequential cycles of polymerization, heat denaturation, and reannealing. |
| Pseudodiploid | Diploid number of chromosomes (46) accompanied by structural rearrangement. |
| Recurrent abnormality |
|
| Telomeric probe |
|
| Translocation |
|
| Tumor suppressor gene |
|
| Whole chromosome painting probe (WCP) | Spans the entire length of chromosomal DNA sequences and, as the name implies, targets the entire length of DNA sequences. |
| Nomenclature | |
| p | = Short arm |
| q | = Long arm |
| + | When placed before the chromosome, denotes a gain of a whole chromosome (e.g., +8) |
| − | When placed before the chromosome, indicates a loss of a whole chromosome (e.g., −7); in rare situations, when placed after the chromosome, as in 5q−, indicates loss of a part of the long arms of chromosome 5 |
| t | translocation |
| del | deletion |
| der | derivative |
| inv | inversion |
| i | isochromosome |
| mar | marker chromosome |
| con | connected |
| nuc ish | nuclear in situ hybridization |
| nuc ish 21q22 (D21S65X2) | two copies of D21S65 DNA segment on chromosome 21 |
| nuc ish 9q34 ( ABL1 x2), 22q11.2 ( BCRx2 ) ( ABL1 con BCRx1 ) | two ABL and two BCR loci, but one of each locus is juxtaposed on one chromosome as a result of t(9;22) |
Fig. 57.1 shows the current cytogenomic methods and their resolution in detecting clonal chromosomal, gene, or other genomic rearrangements and abnormalities in hematologic malignancies.
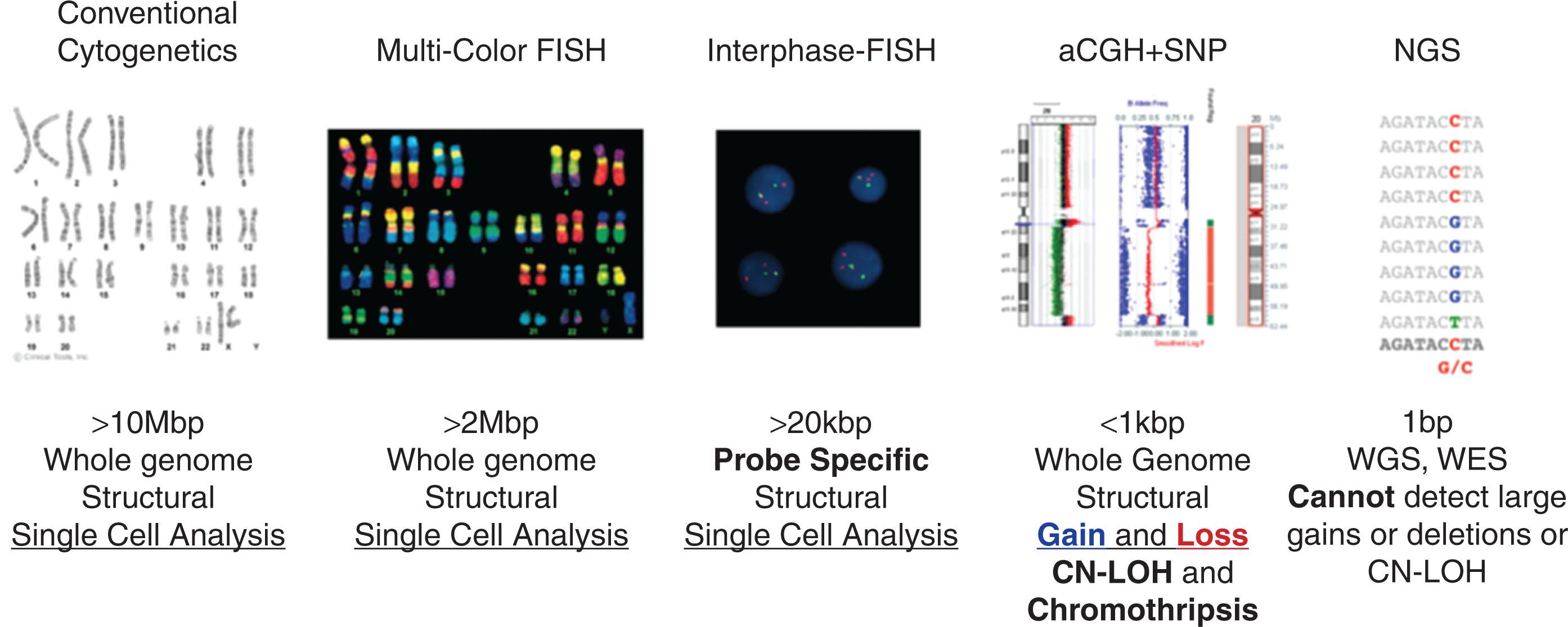
Cells arrested in metaphase are obtained by exposing marrow cells sequentially to mitotic inhibitors, hypotonic potassium chloride, and fixative. Chromosomes obtained from leukemic marrow are then subjected to trypsin-Giemsa banding ( Fig. 57.2 ). The criteria used to define clonal abnormalities are listed in Table 57.1 and described in the International System for Human Cytogenetic Nomenclature, 2021. Table 57.2 describes the optimal logarithm and evolving new testing strategies. These technologies are currently either stand alone or are used in combination.
| Disease | Test | Requirement | Suggested Methodology |
|---|---|---|---|
| CML | Karyotype | Mandatory | Chromosome banding |
| BCR-ABLI gene fusion | Mandatory | FISH or molecular methods/NGS | |
| ABLI mutation when resistance to therapy | Mandatory | Molecular methods | |
| MPN | JAK2, CALR, MPL mutations depending on referral reason | Mandatory | Molecular methods |
| Karyotype, if normal | Highly recommended | Chromosome banding/aCGH+SNP | |
| Myeloid/lymphoid neoplasms with eosinophilia | Recurrent gene fusions involving PDGFRA, PDGFRB, FGFR1, PCM1-JAK2 | Strongly recommended for most patients | FISH, molecular methods |
| Karyotype | Recommended in absence of recurrent gene fusion | Chromosome banding | |
| MDS | Karyotype | Mandatory | Chromosome banding |
| Targeted chromosome abnormalities −5/del(5q),−7/del(7q), MECOM (extended panel +8,del 20q, de( TP53 ) | Recommended | FISH/aCGH+SNP | |
| High resolution chromosome analysis | Recommended | SNP array | |
| Mutation analysis of candidate genes | Recommended | Molecular methods, NGS | |
| AML | Karyotype | Mandatory | Chromosome banding |
| Gene mutations: NMP1,CEBPA, RUNX1, FLT3,TP53, ASXL1 | Mandatory | Molecular methods | |
| Recurrent gene fusions: PML-RARA, CBFB-MYH11, RUNX1- RUNXT1 . Gene rearrangements of KMT2A , MECOM, TP53 | Mandatory | FISH or molecular methods | |
| Treatment-related MDS/AML | Karyotype/FISH: −5/del(5q),−7/del(7q), KMT2A , RUNX1 | Mandatory | Chromosome banding/FISH |
| ALL | Karyotype | Mandatory | Chromosome banding |
| Recurrent gene fusions | Strongly Recommended | FISH | |
| Infants: K MT2A; Pediatric: ETV1-RUNX1, BCR-ABL, TCF3 Adults: KMT2A, ETV1-RUNX1, BCR-ABL1 , | |||
| Hyperdiploidy | Recommended in pediatric | aCGH+SNP | |
| Recurrent microdeletions/AMP/9p21, RUNX1 | Mandatory | FISH/aCGH+SNP | |
| CLL | Deletion 13q14, ATM, TP53 , trisomy 12 | Mandatory | FISH, aCGH+SNP or molecular methods |
| TP53 mutation/IGHV mutational status | Mandatory | Molecular methods | |
| Karyotype | Recommended for clinical trials | Chromosome banding | |
| Multiple myeloma | t(4;14), t(14;16), deletion TP53 gain 1q/del(1p) | Recommended | FISH for gene rearrangements |
| t(11;14), t(14;20), ploidy status (extended panel) | FISH or Array, MLPA for copy number gains and losses | ||
| Other mature B-cell neoplasms | Recurrent gene rearrangements depending on differential diagnosis | FISH | |
| MYC rearrangements for prognostic testing/IgH | FISH |
Fluorescence in situ hybridization (FISH) is a molecular method that allows detection of the number, size, and location of DNA and RNA segments within individual cells in a tissue sample. It is based on the ability of single-stranded DNA to anneal to complementary DNA. In hematologic disorders, the target DNA is the marrow or peripheral blood DNA present in interphase cells or the DNA of metaphase chromosomes that is fixed on a microscope slide.
Fig. 57.3 shows four types of FISH probes that are used alone or in combination to determine both numeric and structural rearrangements: (a) centromere enumeration probes, (b) whole chromosome painting probes, and (c) subtelomeric probes, while Fig. 57.4 shows the four FISH probe strategies used in probe design for detection of chromosomal translocations in hematologic malignancies. The first application of FISH technology for detection of chromosomal translocations in hematologic malignancies occurred when the BCR-ABL1 hybrid gene was identified using two-color FISH in interphase cells as well as in metaphase marrow-derived CML cells. In the standard strategy for interphase evaluation of chromosomal translocation, a DNA probe comprising sequences mapped proximal to the breakpoint in one of the chromosomes involved in reciprocal translocations is combined with a differentially labeled DNA probe that includes sequences mapped distal to the breakpoint in the other chromosome. Nuclei positive for the translocation display one dual-color fusion signal, representing one of the derivative chromosomes generated by the translocation, and two single-color signals, one for each of the normal alleles. This standard FISH strategy has been used for detection of translocations in hematologic disorders at diagnosis.
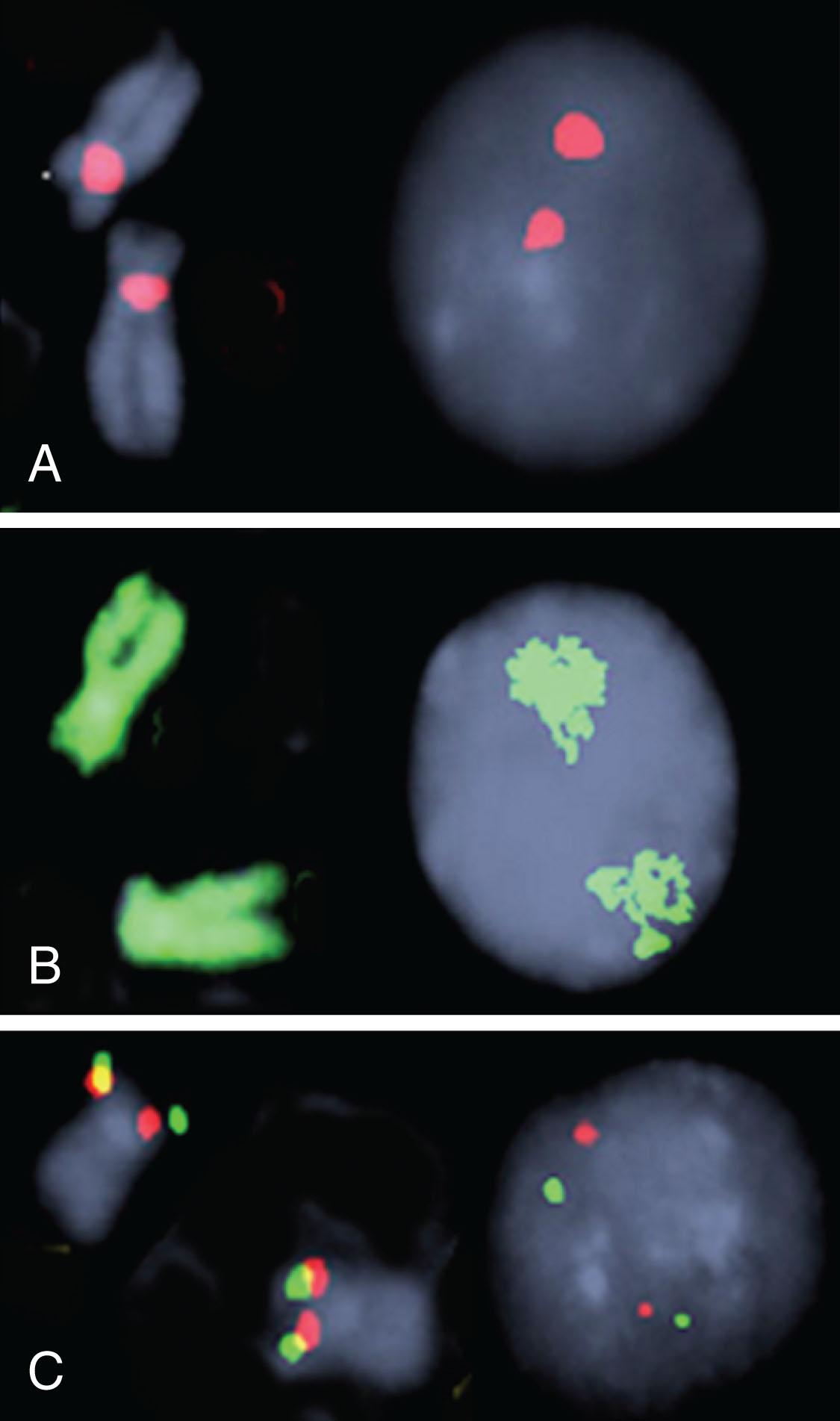
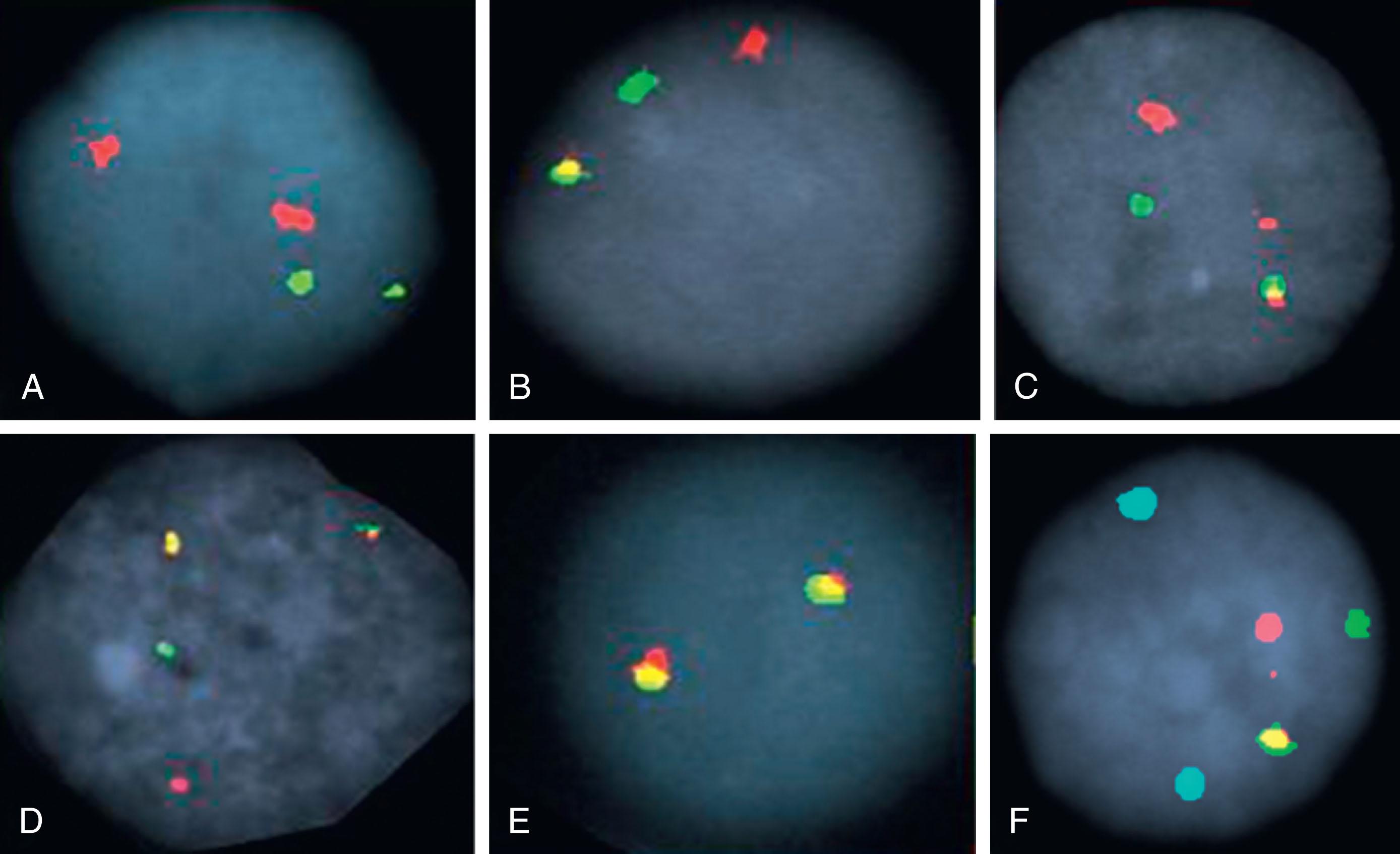
One of the most significant advances in diagnostic leukemia cytogenetics has been the application of interphase FISH. Interphase cytogenetics is the term used to describe detection of chromosomal abnormalities in non-dividing, interphase nuclei ( Fig. 57.5 ). Five aspects of interphase FISH are particularly useful: (1) Interphase cytogenetics allows screening of a large number of cells. This permits investigation of hematologic malignancies with a low mitotic yield, such as chronic lymphocytic leukemia (CLL) and multiple myeloma (MM). (2) Interphase FISH permits detection of chromosomal rearrangements in peripheral blood samples, thus obviating the need for marrow aspiration. For instance, in CML, which rarely yields a large number of dividing cells in peripheral blood, conventional cytogenetics usually is uninformative. However, detection of BCR-ABL1 , the molecular equivalent of the Philadelphia chromosome (Ph), in peripheral blood using interphase FISH provides reliable, fast, quantitative results. (3) Interphase FISH offers a quantitative assay for monitoring disease progression or detection of minimal residual disease (MRD) following chemotherapy or hematopoietic stem cell transplantation (HSCT). (4) Use of specific probe sets allows detection of specific disease-associated abnormalities such as t(8;21), which denotes the M2 subtype of acute myeloid leukemia (AML), or t(15;17), which is associated with acute promyelocytic leukemia (APL), within 4 hours, allowing for timely and appropriate therapy. (5) FISH nomenclature is described in the International System for Human Cytogenetic Nomenclature.
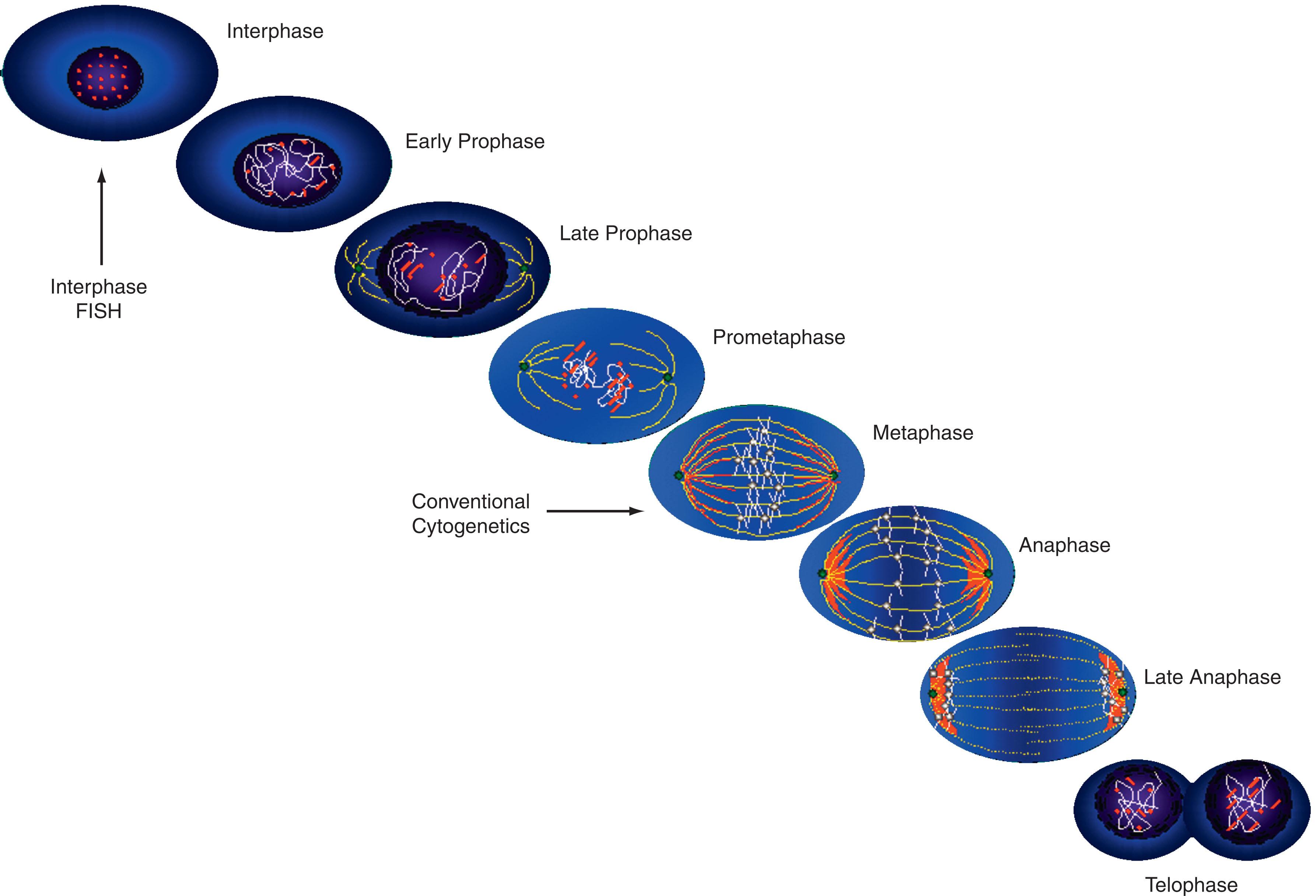
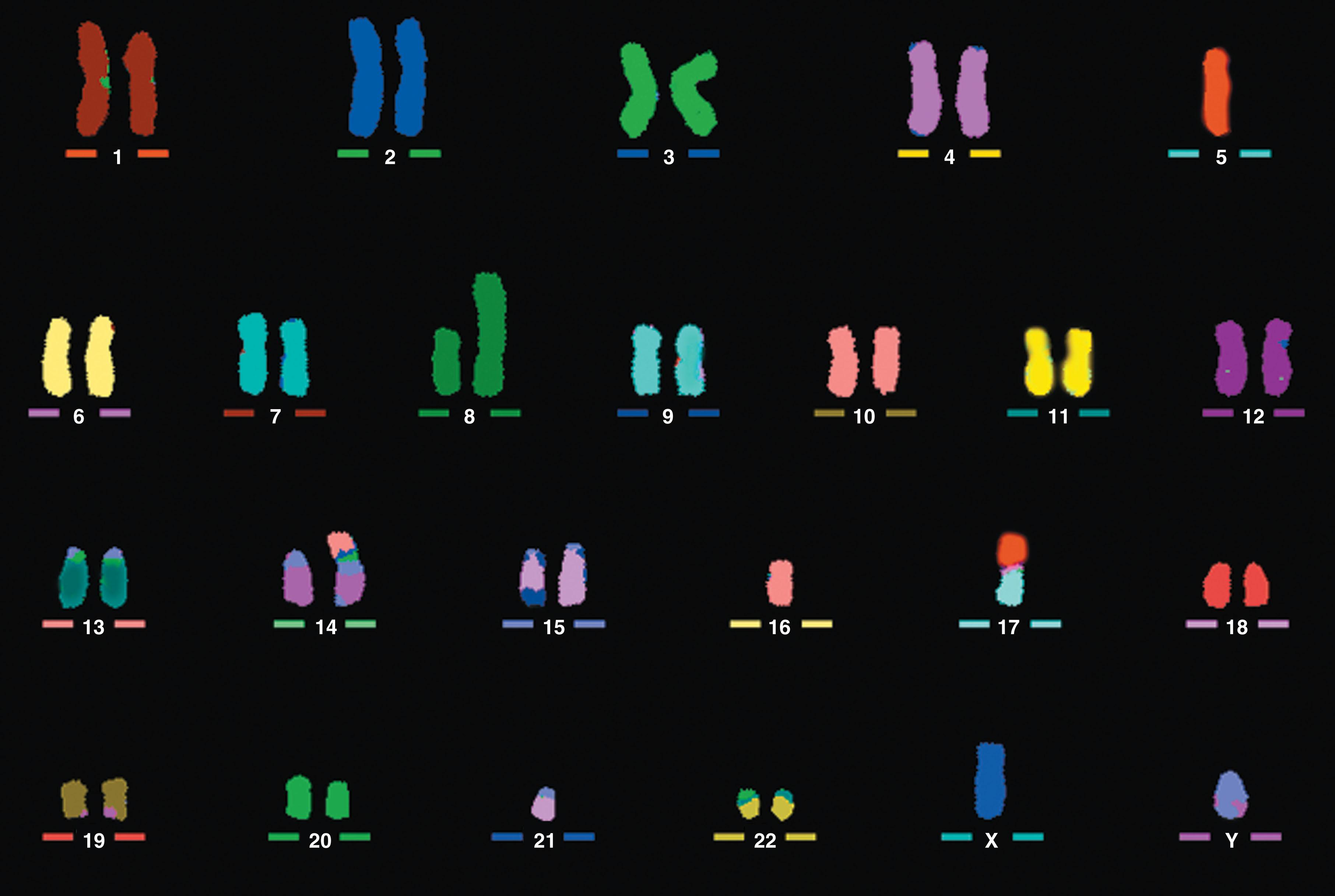
Multicolor karyotyping permits examination of the entire genome in a single analysis ( Figs. 57.1 and 57.6 ). In 1996 it became possible to identify 24 different human chromosomes (22 autosomes and the X and Y sex chromosomes), each with a unique color, with the help of fluorochrome-specific optical filters. This method is called multicolor FISH (M-FISH) . When interferometer-based spectral imaging is used, the method is called spectral karyotyping . The starting point in both methodologies is the use of whole chromosome painting probes for each chromosome. Thus each chromosome is labeled with a different combination of fluorescent dyes and images are sequentially obtained using five different fluorochrome-specific optical filters. A computer program combines the data and displays each chromosome as if it were stained with a distinct color. Spectral karyotyping is based on the use of an interferometer (used by astronomers to measure the light spectra of distant stars) to determine the full spectrum of light emitted by each stained chromosome. A computer program then displays all the chromosomes simultaneously, each with its own unique color. These methods are applied with increasing frequency to resolve complex karyotypes and to resolve the origin of the marker chromosome.
Another powerful method used for identifying the location of chromosomal gains, losses, deletions, or amplifications, without prior knowledge of the chromosomal target that may be altered, is comparative genomic hybridization (CGH) (see Fig. 57.1 ). Briefly, isolated DNA from a patient’s marrow or tumor tissue is labeled with a one-color fluorochrome (e.g., red), whereas DNA isolated from normal control tissue is labeled with a different color (e.g., green). These differently labeled DNAs are hybridized against each other in a competitive hybridization reaction onto normal metaphase spreads. Computer-assisted image analysis detects colors generated after hybridization, which indicate equal hybridization, relative excess, or deficiency of the target DNA (relative to control). The ratio of color intensity provides a “copy number” karyotype.
A particularly useful investigational approach is a “microchip array” in which labeled DNA or RNA from the sample of interest is hybridized with defined target sequences immobilized on a solid support system. This method can screen for genes that are gained/amplified or deleted from the genome on a large scale or, in the case of RNA, to learn whether such genes are expressed at a particular stage of disease. The first example of successful application of the RNA microchip array technique was the discrimination of AML from acute lymphoblastic leukemia (ALL) based solely on gene expression. As shown in Fig. 57.1 , in the array CGH (aCGH) procedure, large-insert genomic clones, oligonucleotides, or single-nucleotide polymorphisms (SNPs) have replaced metaphase chromosomes used in routine CGH. Array CGH is a higher-resolution CGH technology of approximately 5 to 50 kb and provides diagnostic information for diseases associated with DNA dosage. It can also be used to discover previously unexpected sites of altered gene dosage associated with specific hematologic malignancies. The concept of obtaining gene copy number from multiple genome locations in a single measurement has been used to characterize numerous hematologic malignancies over the last 17 years, and its clinical utility is demonstrated throughout this chapter. Nowadays, SNP arrays are also used to genotype a few hundred to millions of SNPs in order to detect rare and common genomic rearrangements (see Fig. 57.1 ). These arrays require hybridization of only the test sample onto the array, unlike aCGH, which relies on co-hybridization of test and reference DNA. At this time aCGH+SNP arrays are used together in one study and they provide exon-level resolution of genomic changes.
The most advanced genomic technologies currently known is next generation sequencing ( Chapter 3 ) or NGS (see Fig. 57.1 ). These approaches use a range of techniques that enable sequencing of hundreds of thousands of nucleic acids simultaneously. In order of complexity, these approaches include sequencing of gene panels, exome sequencing (protein-coding genes), transcriptome (expressed RNA), and sequencing the whole-genome sequencing (WGS). Implementation of these techniques requires several arrays of several hundred thousand sequencing templates in parallel generating up to several hundred million short reads of DNA sequence per lane. The basic principle of NGS involves a process of DNA fragmentation, adapter ligation, and immobilization of the fragments via the adapters to create libraries. The libraries then undergo a process of amplification, generating multiple copies of each DNA fragment which are then sequenced in parallel by a fluorescence- or chemiluminescence-based method, yielding billions of short sequence reads ( Chapter 3 ). These reads are then aligned to the human reference genome and highly efficient algorithms are used to map these complex genomes. Because of its cost effectiveness and enormous sequencing capacity, NGS is revolutionizing hematologic malignancy research by facilitating the discovery of disease-initiating mutations, identifying novel drug targets, and allowing for the first time personalized treatment strategies for hematologic malignancies. Exome sequencing is a relatively inexpensive approach to identify protein-coding mutations, but is incapable of identifying structural genetic rearrangements, deletions, and insertions of DNA, a hallmark of many hematologic malignancies. Exome sequencing is performed at a depth of 100- to 200-fold coverage of the haploid genome, which enables detection of mutations present in leukemic subclones, which is important in the study of relapse. Transcriptome sequencing involves sequencing of genes actively expressed and can be adjusted to selectively study RNA transcripts that encode proteins (mRNA), transcripts regardless of coding potential (RNA), or a variety of small and noncoding RNA transcripts. RNA sequencing is a highly informative approach which enables identification of chromosomal rearrangements that result in the expression of chimeric fusion genes and sequence mutation detection. WGS usually involves sequencing leukemic and nonleukemic DNA from an individual. Although it is the most comprehensive modality, WGS may not identify all genetic alterations caused by variations in sequence coverage and difficulties in sequencing complex and GC-rich regions of the genome (including gene promoters). These modern cytogenomic methods have increased the resolution at which chromosomal and gene rearrangements can be identified. Conventional cytogenetics, FISH, and aCGH+SNP along with NGS are complementary. Each has its own pros and cons in investigating genomic rearrangements of malignant cells.
Although conventional cytogenetics is the comprehensive study of all chromosomes, it requires a large number of dividing cells, which, in some diseases, such as myelofibrosis, is difficult to obtain (see Table 57.2 ). Furthermore, many small deletions or structural rearrangements are beyond the microscopic level of detection. FISH should be used in conjunction with conventional cytogenetics with both interphase and metaphase cells. It is a more sensitive method and detects rearrangements smaller than 1 kb. The main disadvantage of interphase FISH is that it cannot be used unless a known abnormality is suspected. When the abnormality is known, interphase FISH identifies the clonal aberration at the single-cell level. aCGH+SNP provides genome-wide higher resolution analysis and important information concerning genomic changes in patients with a normal karyotype as well as acquired loss of heterozygosity, and chromothripsis (chromosome shattering), which are important prognostic and predictive tools. With the introduction of high throughput genomic technologies such as NGS, FISH-based chromosome level detection has gradually changed focus to genome-wide detection of single nucleotide and copy number variants that are common in leukemia. It is evident that identification of chromosomal aberrations by molecular cytogenomic techniques is important in detecting novel chromosomal rearrangements and genes involved in leukemogenesis. Understanding the basis of these techniques and their application is critical in the accurate diagnosis of hematologic malignancies. Table 57.2 describes the optimal algorithm and evolving new testing strategies. New technologies, capable of simultaneously detecting copy number changes, structural variants, and mutations, are expected to be used in future in a diagnostic setting.
Genome sequencing has rapidly become routine practice. However, the intimate relationship between DNA sequencing, chromosome structure, and the position of chromosomes in the nucleus is not fully understood. The term “ chromosomics ” was recently proposed in order to integrate the original definition of cytogenetics (chromosomes and cytology) with genomics (gene content, structure and function) to ensure a closer integration of these fields. This term chromosomics is encompassing the integration of the latest advances in cytogenetics, genome sequencing, epigenomics, and cell biology. Chromosomics approaches in future will lead to additional success in answering fundamental biological questions in hematologic malignancies.
In early 2021 three studies used novel next-generation karyotyping which employed WGS as a potential replacement for conventional cytogenetics ( Chapter 3 ). These initial studies focused on patients with AML and myelodysplastic syndrome (MDS) and the genetic profiles and sequencing approaches were tested against the backbone of orthogonal results obtained at the same time using cytogenetics, FISH, and polymerase chain reaction (PCR) and/or NGS for detection of acquired somatic mutation. In a study reported by the Washington University group, in 235 patients who had undergone successful cytogenetic analysis, WGS provided rapid and accurate genomic profiling in patients with AML or MDS after modifications of sample preparation, sequencing, and analysis to detect mutations to be used for risk stratification using existing European Leukemia Network (ELN) guidelines. The WGS detected all 40 recurrent translocations and 91 copy-number alterations that had been identified by cytogenetic analysis. In addition, next-generation karyotyping identified new previously unrecognized genomic events in 17% of patients. In 117 consecutive patients analyzed prospectively WGS provided results within a median of 5 days and additional genetic information was identified in almost 25%, which changed the ELN and Revised International Prognostic Scoring System (IPSS-R) genetic prognostic risk categories in 16% of patients. Standard AML risk groups, as defined by sequencing results instead of cytogenetic analysis, correlated with clinical outcomes. WGS was also used to stratify patients who had inconclusive results by cytogenetic analysis into risk groups in which clinical outcomes were measurably different. These initial data are highly promising because they provided a greater diagnostic yield than conventional cytogenetic analysis and allowed for more efficient risk stratification on the basis of standard risk categories. Moreover, the speed by which the clinically relevant genomic profiles were obtained allowed reports to be generated in as little as 3 days.
The question of whether cell proliferation is monoclonal or polyclonal is fundamental to understanding the underlying origins of hematologic malignancies. Markers of clonality are used to determine the origin of disease; to differentiate malignant from nonmalignant populations; to establish hematopoietic hierarchy, clonal evolution, and clonal remission; and to delineate steps involved in the multistep pathogenesis of hematologic malignancies.
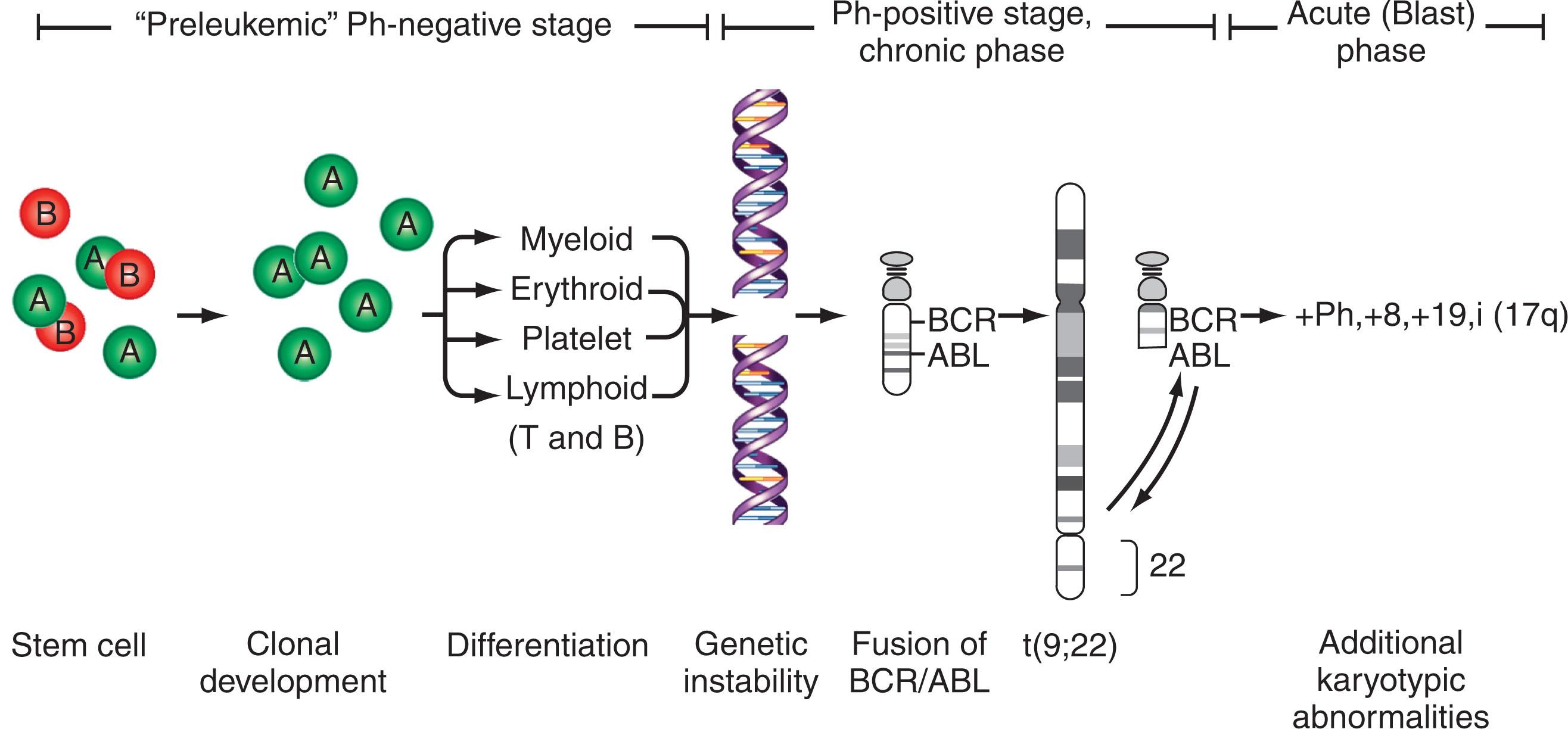
The clonal origin of leukemias and lymphomas can be assessed by either intrinsic or extrinsic cellular markers. Intrinsic cellular markers are specific for a cell population, arising either during normal differentiation or as a part of disease process. For instance, cell surface-associated immunoglobulin (Ig) markers such as the λ or κ light chain or idiotypes and T-cell receptors (TCRs) can be useful for evaluating lymphoid malignancies. Application of IgH markers demonstrated for the first time that MM was of clonal origin. Somatic cytogenetic alterations are useful intrinsic markers for identifying abnormal clones and following disease progression. Thus the observation of identical chromosome anomalies in different cells of the same tumor is evidence of clonality. Since the discovery of the Ph in 1960 it has been well established that nonrandom, recurrent chromosomal abnormalities characterize many hematologic malignancies. The finding of the Ph in different CML-derived hematopoietic cell lineages led to the hypothesis that CML originates in a single precursor cell that has a clonal development pattern. Moreover, the presence of additional recurrent chromosomal abnormalities in the Ph-positive clone (such as trisomy 8, duplication of the Ph, or trisomy 19) not only indicates the progression of the disease to accelerated phase or blast crisis, but also demonstrates the subclonal evolution of the Ph-positive clone. Currently, disease-associated somatic genomic mutations, such as rearrangements of KMT2A (MLL) , runt-related transcription factor gene (RUNX1), ETV6, PML-RARA , and many others, can be identified by PCR-based assays, FISH assay, and novel aCGH and NGS, and may serve, with or without conventional cytogenetics, as intrinsic markers of disease processes.
On the other hand, extrinsic marker systems use cellular mosaicism that is completely independent of the disease being studied and is not restricted to the cell lineages. Individuals with Turner or Klinefelter syndrome are mosaic for XX or XY and monosomy X cells or XXY and XY cells, respectively. The mosaicism created by X-chromosome inactivation in females is much more widely applicable, and initially provided fundamental insights into the pathogenesis of hematologic malignancies. Original studies with X-linked glucose-6-phosphate dehydrogenase ( G6PD ) as a marker of clonality were based on the Lyon hypothesis, which asserts that early in embryogenesis, one X chromosome in females is inactivated in somatic cells and the activation status is stably transmitted to daughter cells during mitosis ( Fig. 57.7 ). The choice of maternal versus paternal X-chromosome inactivation is random; however, once it occurs, it is maintained in all daughter cells. Random X inactivation occurs by embryonic day 6.5 around the start of gastrulation and results in a mosaic pattern that characterizes adult females. Therefore an adult female is a mosaic for two-cell populations, one expressing genes from an active X chromosome and the other expressing genes from the inactive X chromosome. Incidentally, mammalian X-chromosome inactivation is a mechanism that equalizes the dosage of X-linked genes between sexes. Although the exact mechanism of X-chromosome inactivation remains to be elucidated, the process of X inactivation starts with methylation of CpG islands. The inactivation process is believed to occur before differentiation of the embryonic stem cell into various cell lineages. Hematopoietic cells do not originate from a single embryonic stem cell but from several hematopoietic stem cells, thereby allowing for mosaic expression from both X chromosomes.
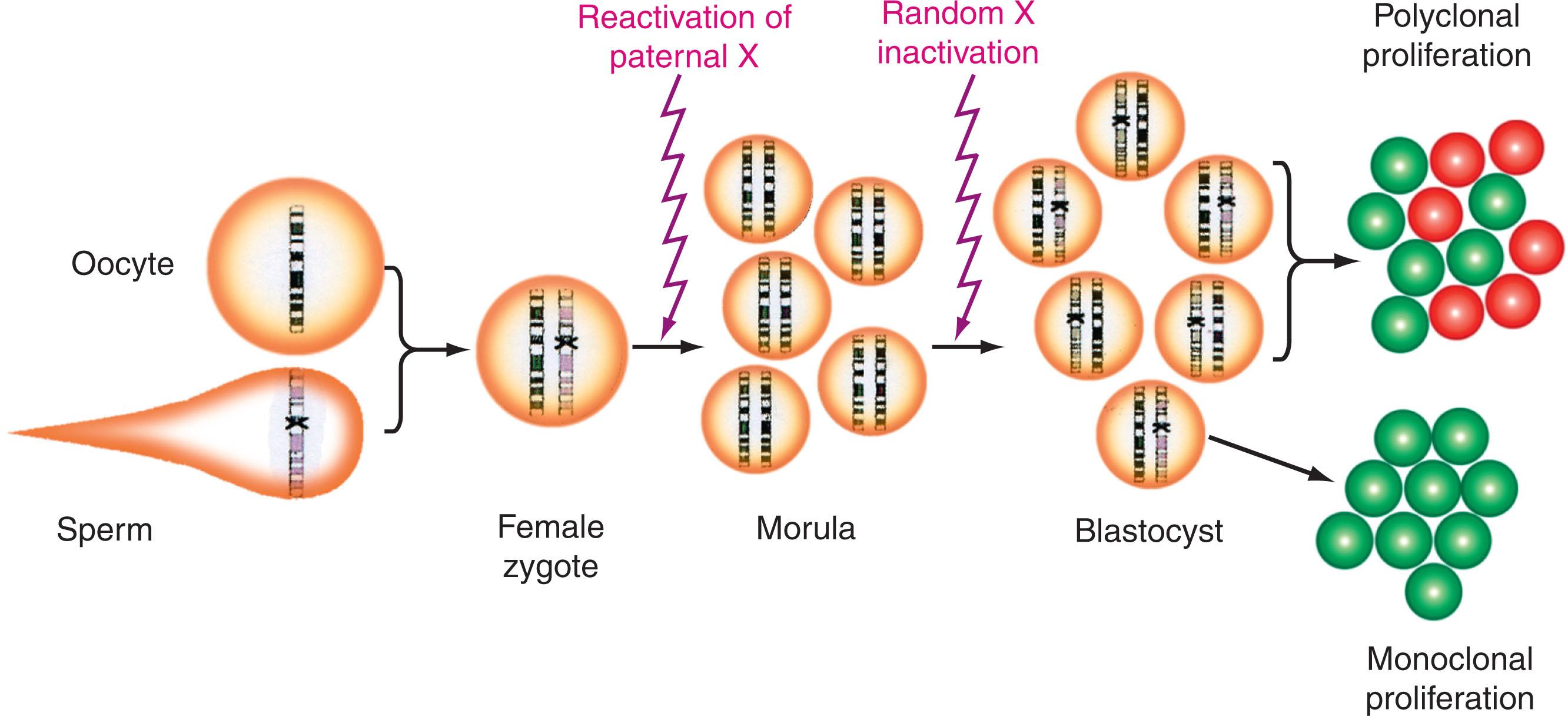
The observation that human females were heterozygous for the G6PD variant A and A − and that two mosaic cell populations may be distinguishable by electrophoretic mobility was reported in the 1960s. The X-inactivation G6PD mosaic system was then applied to the study of clonality in human tumors (uterine leiomyomas) in 1964 by Gartler and Linden. In females who were heterozygous for the G6PD polymorphism and had malignant hematologic disorders such as CML, the finding of a single G6PD type in marrow or blood cells and both the A and B type G6PD in tissues not involved by the malignant process first demonstrated that CML was of clonal origin and provided evidence that the malignant transformation occurred at the level of a stem cell common to most hematopoietic cell lineages. Additional studies with heterozygous G6PD females who had CML demonstrated that some CML-derived B lymphocytes had a single G6PD type, but these clonal cells were Ph-negative; thus leukemic transformation might predate development of the chromosomal abnormality. This observation provided evidence that CML has a multistep pathogenesis. Application of this approach to hematologic malignancies demonstrated the clonal and stem cell origin for AML, ALL, Ph-negative myeloproliferative neoplasm (MPN), MDS, and CLL. Studies using G6PD were particularly useful in the investigation of red blood cells and platelets in hematologic malignancies because the absence of nuclei in these cells did not allow them to be studied with cytogenetics or DNA analysis. Although it is now considered common knowledge that hematologic malignancies are characterized by clonal development, this concept was largely developed by Dr Phillip Fialkow.
Despite the importance of the G6PD approach, it is limited by the rarity of females who are heterozygous for the G6PD isoenzymes. An alternative and more extensive DNA-based X-chromosome clonal assay used common polymorphic markers that are caused by changes in DNA methylation patterns that accompany inactivation of the X chromosome. These X-linked loci such as phosphoglycerine kinase, hypoxanthine phosphoribosyltransferase, DXS25 (M27β), and human androgen receptor (HUMARA), have been extensively used in assessment of clonality, and can be used to identify clonal cell populations in virtually all females. DNA-based marker systems rely on a sequence polymorphism that has adjacent differences in methylation on the active and inactive X chromosomes. The inactive X chromosome is more highly methylated than its active homologue, but this is only true for certain regions of genes as 10% to 20% of X-linked genes escape inactivation and can be found both in clusters and in isolation. The most widely used HUMARA assay appears to maintain stringent methylation differences. The number of CAG tandem repeats differentiates the maternal from the paternal X chromosome.
The utility of the DNA-based X-chromosome clonal assay is limited to females younger than 60 years because they usually have 1:1 distribution of two-mosaic–cell population. A ratio greater than 3:1 is found in women older than 60 years, probably as a result of stem cell kinetics influenced by X-linked genetic factors. When the ratio of two-cell populations is greater than 3:1, this phenomenon is called a skewed X-inactivation pattern . With the HUMARA assay, acquired unequal or skewed X-chromosome inactivation (excessive lyonization) is found in 35% to 40% of women older than 60 years. Thus X-chromosome–based clonality studies must incorporate age-matched controls. More recently acquired somatic mutations using NGS have been useful in documenting clonal hematologic malignancies.
The observation that monozygotic twins share identical but non-constitutive and clone-specific fusion gene sequences (e.g., ETV6-RUNX1 ) in pediatric ALL provided the first unambiguous evidence (in 2003) that genetic lesions, generated by chromosomal translocation, arise in utero. These studies initiated by Mel Greaves and his colleagues contributed a wealth of knowledge about founder mutations, subclonal development, clonal origin, and the evolution of disease. The initiating lesion and premalignant clone is shared by the twins as a consequence of intraplacental vascular anastomoses and blood cell chimerism. The twin data were endorsed by backtracking of prenatal-initiating genetic lesions in the archived blood spots, or Guthrie cards, of patients with ALL. These data were interpreted to suggest that ETV6-RUNX1 was likely a critical initiating lesion for ETV6-RUNX1 -positive ALL. However, such fusions were detectable in cord blood from newborn infants at rates approximately 100-fold higher than the incidence of ALL, suggesting an obligatory requirement for additional mutations for leukemia development to occur. Over a period of 10 years these results were confirmed and the in utero origin of MLL , BCR-ABL1 , and RUNX1-RUNXT1 fusion rearrangements further documented, providing direct evidence for a prenatal origin of many childhood leukemias.
Results from genome sequencing of ETV6-RUNX1 fusion region suggests it arises as a consequence of nonhomologous end-joining in the pro-B-cell stage with possible self-renewal capacity to downstream B-cell precursors. Additional evidence was provided by screening for trisomies in stored cord blood and the data indicated that ∼6% of enriched CD34+/CD19+ B progenitor cells carry trisomies frequently seen in hyperdiploid childhood ALL. Using novel SNP as well as other technologies, it has become apparent that ALL has multiple genome copy number variations (CNVs), mostly deletions, and these CNVs are distinctive between a pair of twins, indicating a secondary, postnatal origin.
When genotyping sequencing combined with other molecular technologies of five pairs of monozygotic twins with concordant ETV6-RUNX1 -positive ALL, Greaves and his colleagues demonstrated that all recurrent CNVs (32 in total) were different within twin pairs providing strong evidence that they are probably secondary events and postnatal in origin in both twins as well as in non-twins with ALL. Another two twin pairs who shared a monochorionic placenta and had Ph-positive ALL, were also studied by Greaves and his colleagues. These studies provided confirmation of the previous observation that BCR-ABL1 is not sufficient to cause Ph-positive leukemia. Twin A presented with ALL at age 3.8 years and twin B presented at age 4.1 years. Both had an identical BCR-ABL1 fusion transcript and both received an allogeneic stem cell transplantation from the same matched sibling donor but 7 months later twin B died, whereas twin A remained in good health 8 years following the transplantation. SNP analysis revealed that twin A had a subclone with trisomies for chromosomes 4, 6, 9, 14, 17, and X, tetrasomy 21, and a gain of 22q11.1–q11.23 region, whereas twin B had deletions of EBF1 and IKZ1 . These results provide direct evidence that rearrangements additional to BCR-ABL1 are postnatal in origin, they represent subclonal evolution, and BCR-ABL1 by itself is not sufficient for development of Ph-positive ALL. Recently two groups have reported, using mathematical models, that some patients’ MPNs acquire the JAK2 mutation in utero.
These mutation-driven natural history studies provide evidence for sequential multistep pathogenesis of hematologic malignancies with sequential accumulation of genetic changes, which may be linear through clonal succession but that clonal evolution in most leukemias is complex and may represent a branching structure, as predicted by Darwin.
High-throughput sequencing has provided novel observations which have added to our current understanding of the initial mutations that occur in premalignant stem cells and their clonal development that eventually leads to myeloid malignancies ( Chapter 157 ).
Clonal mosaicism for large chromosomal anomalies (duplications, deletions, and copy number neutral (CN-LOH)), using SNP microarray data from 50,000 subjects in the GENEVA study, indicated that CH is infrequent (<0.5%) from birth until 50 years of age after which its frequency rapidly increases to 2% to 3% of the elderly. It has been estimated that individuals with clonal hematopoiesis (CH) have 10-fold higher risk of developing a subsequent hematologic malignancy. These age-related mutations were confirmed by analysis of mutation acquisition in hematopoietic stem cells, over time, through whole-exome sequencing of single hematopoietic stem cell-derived colonies, which showed that the total number of mutations in healthy individual stem cells increases with age. Therefore, the term CH describes the acquisition of somatic mutations in hematopoietic stem and progenitor cells resulting in clonal expansion in individuals with no overt hematologic disease. Moreover, sequencing of multiple elderly women provided evidence of CH based on X inactivation pattern and recurrent somatic mutation in TET2 gene. Subsequent analysis of 182 additional elderly women with CH determined that more than 5% of these individuals had mutations in TET2 ( Chapter 157 ).
Investigation of 82 paired bone marrow and blood samples from carefully selected healthy adult volunteers using a panel of 41 genes known to be mutated in myeloid malignancies, with 1% threshold of detection revealed that bone marrow clones were found in almost 40% of healthy volunteers over 50 years of age. The most frequent mutations included DNMT3A and TET2 . Variant allele frequencies were highly concordant between blood and bone marrow samples.
However, individuals with CH may live for many years and decades without developing hematologic malignancies though they are at increased risk as compared with those without mutations. These studies may explain the higher frequency of myeloid malignancies in the elderly population ( Chapter 157 ).
Loss of Y chromosome (LOY) was first described in 1963 in cultured blood leukocytes as a sole cytogenetic abnormality in bone marrow cells which represents one of the most common acquired, post zygotic somatic genomic alterations in elderly men. Approximately 6% of male bone marrow karyotypes from normal individuals and 16% of bone marrow karyotypes with hematologic malignancies show a LOY. The incidence of LOY in bone marrow cells increases proportionally with aging with up to 20% of healthy men over 80 years of age showing LOY using standard G-banding techniques. Mosaic LOY in blood has been reported to be associated with higher risk of all-cause mortality and overall lower life expectancy. LOY has been described most frequently in patients with myelodysplastic disorders and has been reported in MPN, AML, and other myeloid diseases as well as in various solid tumors. Since MDS and MPN are primarily diseases of elderly individuals it is often difficult to separate age-associated LOY from disease-associated LOY. Although the incidence of LOY in MDS is up to 30%, the sole LOY is lower, ranging from 4% to 10%; the WHO does not recognize LOY by itself to be useful in cytogenetically identifying a case of MDS unlike other abnormalities such as del(5q), del(7q), +8, and del(20q).
Recent observations that a higher LOY burden is associated with a higher likelihood for progression to a myeloid malignancy represents the basis for a proposal to include LOY as another form of CH with striking similarities with CHIP. Among 73 studied male patients with sole LOY and a median age of 75 (range 29 to 90), those who had equal or greater than 75% of marrow metaphase cells with LOY had a significantly greater chance of developing a myeloid neoplasm and a higher lifetime incidence of diagnosis of MDS ( P < .0001). Male individuals with greater than 75% of metaphase cells with LOY also had greater likelihood of having somatic mutations ( P = .0009) and greater numbers of these mutations ( P = .0002). The presence of metaphase cells with equal or less than 25% LOY was associated with normal bone marrow morphology and a lower likelihood of progression to MDS. Multivariate analysis showed that these observations are consistent with samples showing 75% or more metaphase cells with LOY are myeloid disease-associated and not related due to aging. Therefore, it has been proposed that LOY represent a marker of CH and as such should be included in the definitions of CHIP in clinical and research settings.
The relationship between isolated loss of X chromosome (LOX) and aging was first documented in 1995 using interphase FISH method applying to peripheral blood cells of 90 healthy females (age: 1 week to 93 years) with a frequency of 3.2% below 51 years to 5.1% at age 51 to 93 years. This observation was confirmed a few years later in a cohort of 655 females and the frequency of LOX is ranging from 0.07% in patients older than 16 years to 7.3% in individuals older than 65 years. LOX was occasionally reported in BM cells at the frequency of 0.1% of females with the median age of 71.
Acquired trisomy 15 as an isolated abnormality is rarely detected. Approximately 0.1% to 0.2% of elderly patients show +15, frequently in association with LOY. In bone marrow cells, when I-FISH is used, +15 may represent a small clone of up to 24% of nondividing cells but none of these patients developed a myeloid neoplasm after a median follow-up of 28 months (range 0 to 149 months). These observations suggest that +15, like LOY, when present as a small clone is not disease-related but represents an abnormality associated with CH and is most likely age-related.
The World Health Organization (WHO) characterizes MPNs as clonal stem cell disorders. CML has a unique place among hematologic malignancies and is described separately from the other Ph-negative MPNs.
Knowledge of the origins of CML has accumulated over the last 61 years and serves as a classic example of molecular medicine at its best ( Table 57.3 ). The Ph chromosome is the first example of a specific chromosomal abnormality associated with a malignant disease. ABL1 and BCR genes are the first oncogenes localized at the site of a chromosomal breakpoint in t(9;22)(q34;q11.2). The BCR-ABL1 fusion leads to a “hybrid” gene, resulting in the production of a dysregulated tyrosine kinase protein. Finally, imatinib mesylate, a specific tyrosine kinase inhibitor (TKI), was the first rationally designed targeted form of cancer therapy.
| 1960 | Philadelphia chromosome (Ph) is identified. |
| 1973 | Ph is t(9;22)(q34;q11.2). |
| 1983 | ABL1 is translocated from chromosome 9 to chromosome 22. |
| 1984 | BCR is localized to 22q11. |
| 1987 | Ph′ is BCR-ABL1 fusion. |
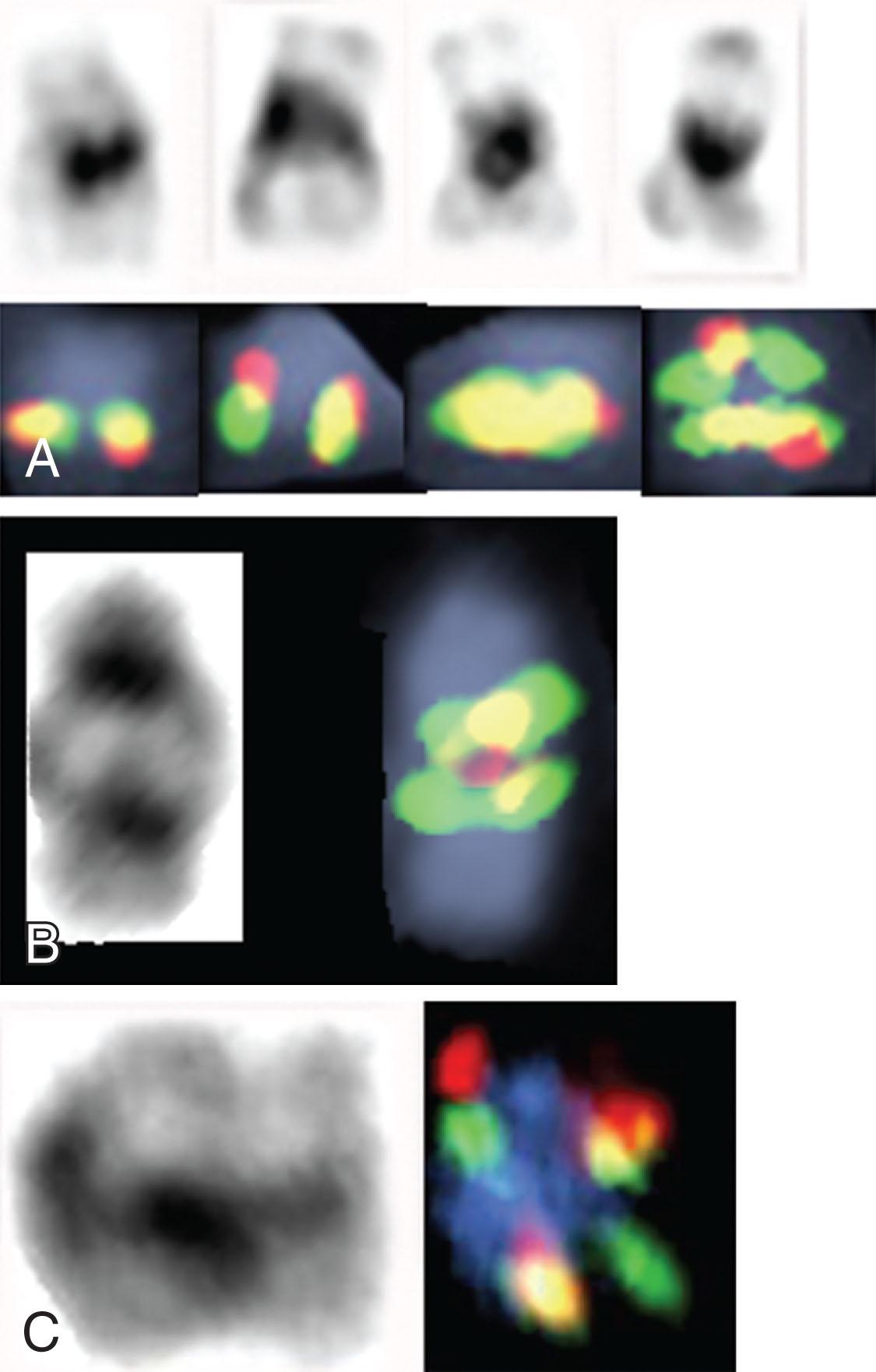
The Ph chromosome, named in honor of Philadelphia, the city of its discovery, was described for the first time in 1960. It represents a signature genomic rearrangement occurring in more than 95% of patients with CML. Approximately 3% of all pediatric leukemias are Ph-positive CML. The incidence in children increases with age and it is exceptionally rare in infants. The Ph chromosome results from a balanced translocation t(9;22)(q34;q11.2) ( Fig. 57.8A–B ). The Ph chromosome arises postzygotically, being found only in hematopoietic tissue. The finding of the Ph chromosome in myeloid cells, erythroid cells, eosinophils, monocytes/macrophages, basophils, and B lymphocytes, along with the absence of the Ph chromosome in cultured marrow fibroblasts, supports the concept that the Ph chromosome results from a specific rearrangement in a multipotent hematopoietic stem cell and that it is an acquired rather than an inherited abnormality. Of interest, the Ph chromosome is rarely identified in T cells. T lymphocytes are long-lived cells and may antedate the development of CML. These observations combined with studies exploiting G6PD heterozygosity provide further evidence for the concept that CML is a clonal disease arising in a stem cell capable of differentiation into all hematopoietic cell lineages.
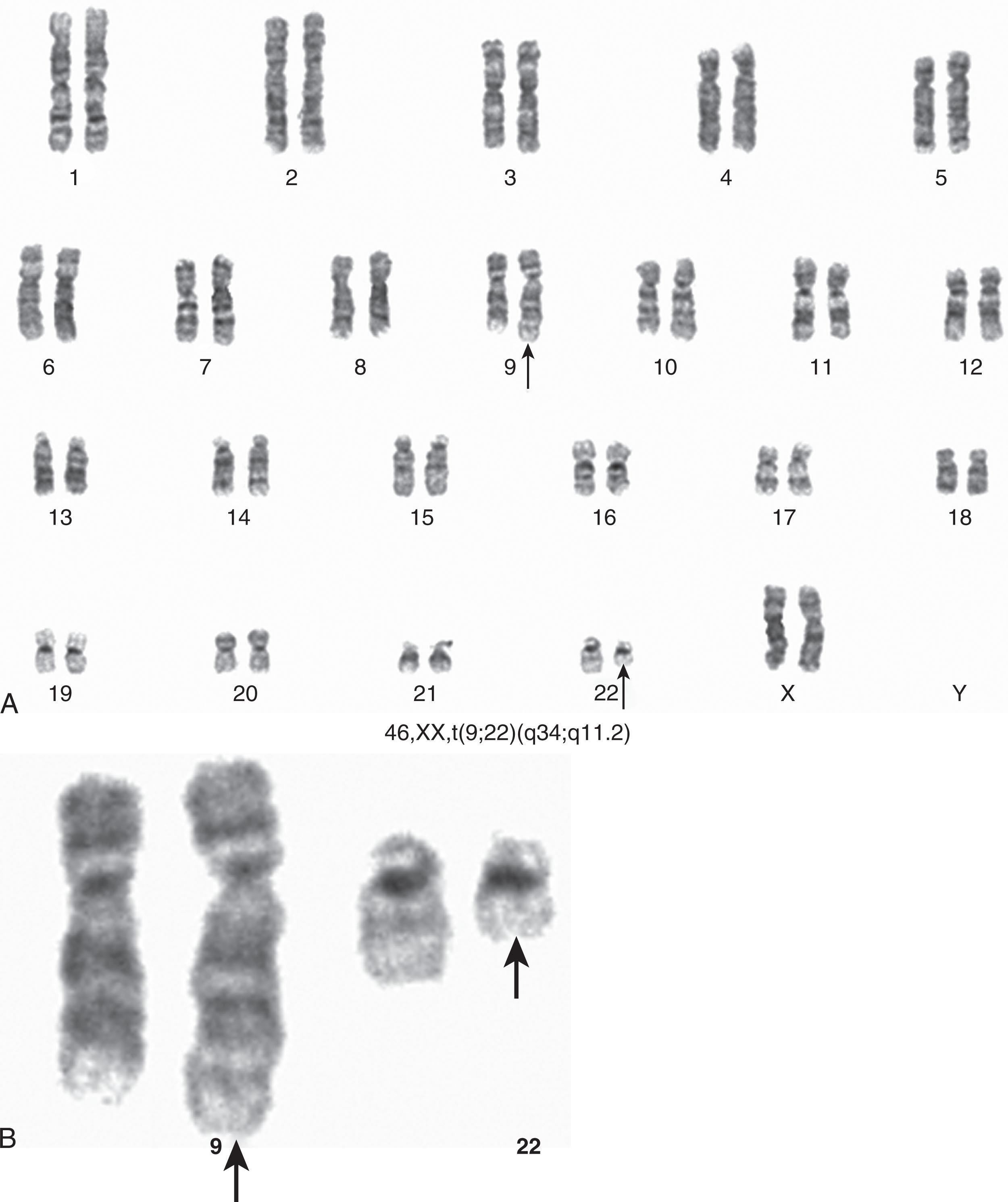
In a review of 1129 Ph-positive patients, the 9;22 translocation was identified in 1036 (92%) cases. Karyotypic analysis of marrow cells in patients with CML is a time-consuming task. However, it demonstrates not only the presence of the Ph chromosome but also the presence of other chromosomal rearrangements (clonal evolution) of clinical significance.
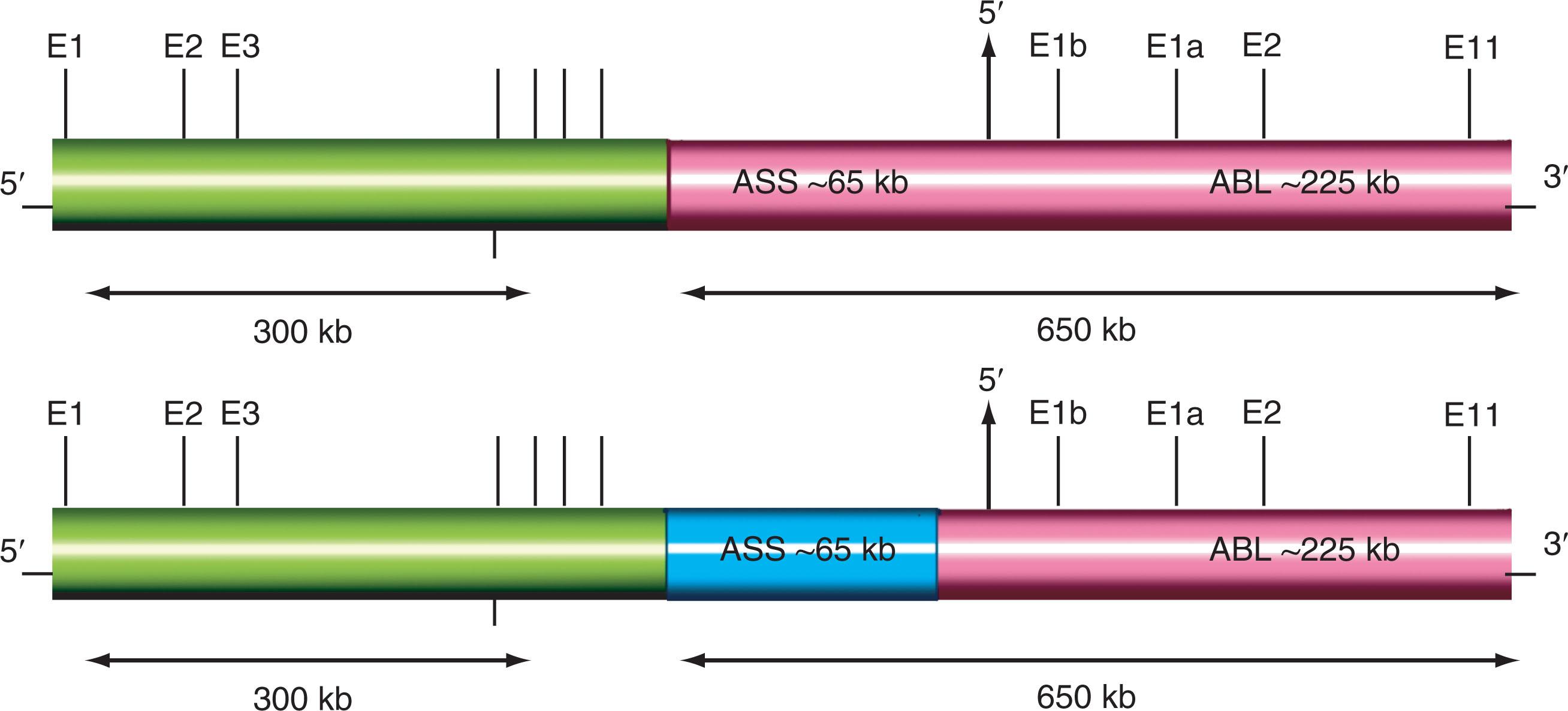
The reciprocal nature of the Ph-positive translocation was confirmed when studies showed that it is the molecular consequence of the translocation of the ABL1 gene from chromosome 9, band region q34, and subsequent fusion to the breakpoint cluster region ( BCR ) gene on chromosome 22, band q11.2 ( Fig. 57.9 ). This creates a hybrid BCR-ABL1 gene that is transcribed into a chimeric BCR-ABL1 messenger RNA (mRNA) and translated into a specific chimeric protein.
Three major breakpoint locations along the BCR gene on chromosome 22 result in three chimeric proteins. They include P210 BCR-ABL1 , P190 BCR-ABL1 , and P230 BCR-ABL1 which are associated with three distinct types of leukemia. P210 BCR-ABL is found in the majority of patients with classic Ph-positive, BCR-ABL1 fusion-positive CML and approximately 30% of patients with Ph-positive ALL. Expression of P190 BCR-ABL1 is seen in 20% to 30% of adults and 80% of children with Ph-positive ALL. Expression of P230 BCR-ABL1 is associated with a rare indolent chronic neutrophilic leukemia variant and up to 1.6% of CML (approximately 50 patients in the worldwide literature have been described). The most common BCR-ABL1 isoform results from a fusion of BCR exon 13 or 14 (e13/e14) with ABL1 exon 2 (a2) producing a fusion gene referred to as e13a2 (b2a2) or e14a2(bra2) producing above-mentioned p210 protein. The coiled-coil domain of the BCR N terminus facilitates dimerization and constitutive autophosphorylation of the ABL1 tyrosine kinase domain resulting in subsequent phosphorylation of numerous substrates including the JAK2/STAT CRKL MAPK and P13K/AKT pathways.
Approximately 1% to 2% of Ph-positive patients with CML express both P210 BCR-ABL1 and P190 BCR-ABL1 and their response to TKI therapy is inferior to patients showing only P210 BCR-ABL1 . The BCR-ABL1 fusion is present in both standard and variant forms, in cases where chromosome 9 involvement is cytogenetically not detectable, and when a masked Ph is present. In the majority of patients, the fusion of ABL1 and BCR takes place on chromosome 22 ( Fig. 57.10A–B ). The BCR-ABL1 fusion transcript is present in neutrophils, monocytes, eosinophils, erythrocytes, B cells, rarely in T cells, and in CD34+ cells and is associated with increased proliferation of CD34+ myeloid progenitor cells but not of other more mature myeloid precursors.
These observations confirm the hypothesis that CML originates in a multipotent stem cell capable of differentiating to all hematopoietic cell lineages with the exception of T cells. These and other studies provide evidence for the existence of clonal BCR-ABL1 fusion-negative stage. The formation of BCR-ABL1 and the Ph chromosome occurs in an already abnormal and genetically unstable clone of pluripotent hematopoietic stem cells. Thus it is the preexisting genetic instability that predisposes to the formation of the Ph chromosome. Once Ph chromosome formation occurs, it confers a further selective growth advantage over normal cells, resulting in overwhelming BCR-ABL1 -positive, Ph-positive marrow cells at the time of diagnosis of CML.
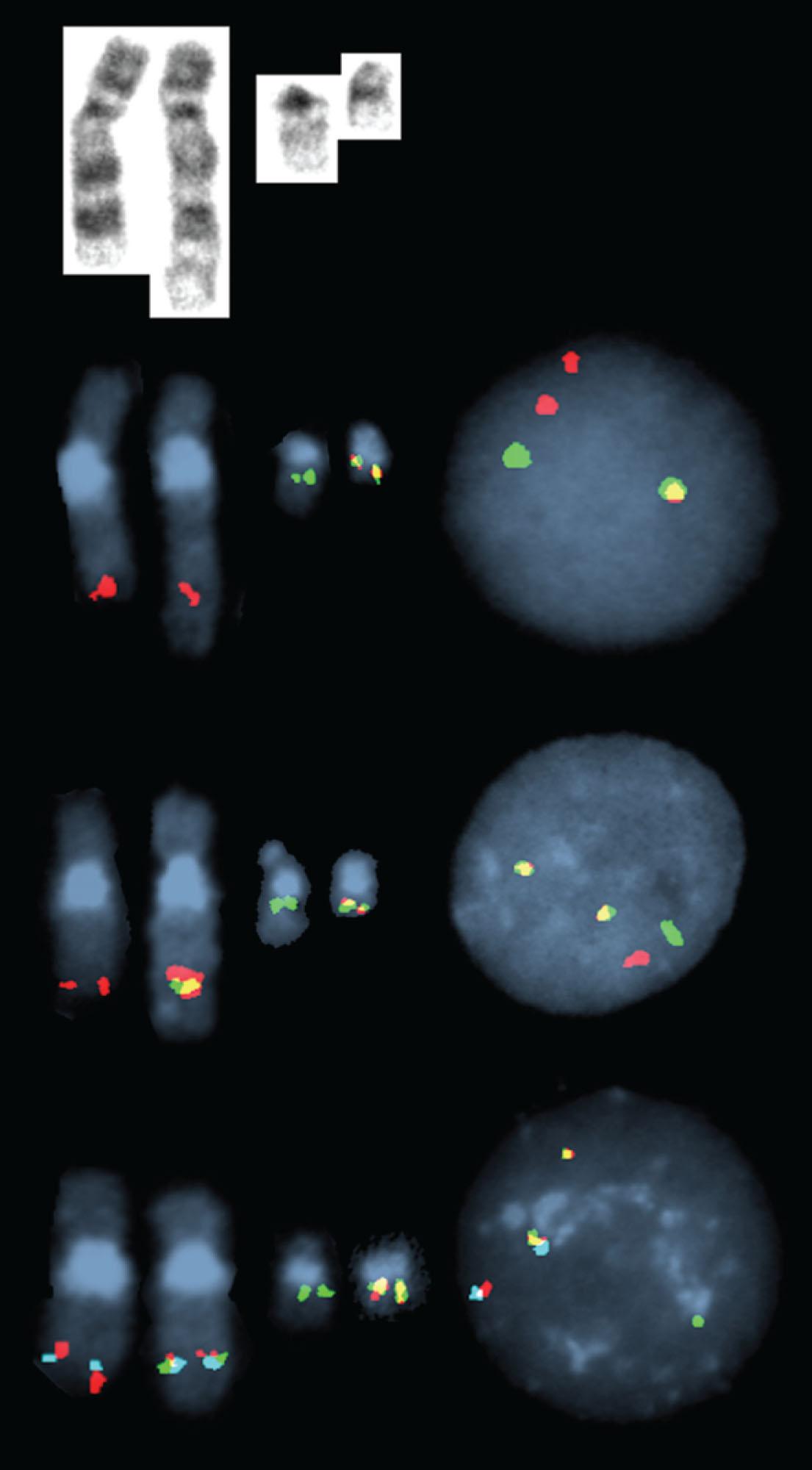
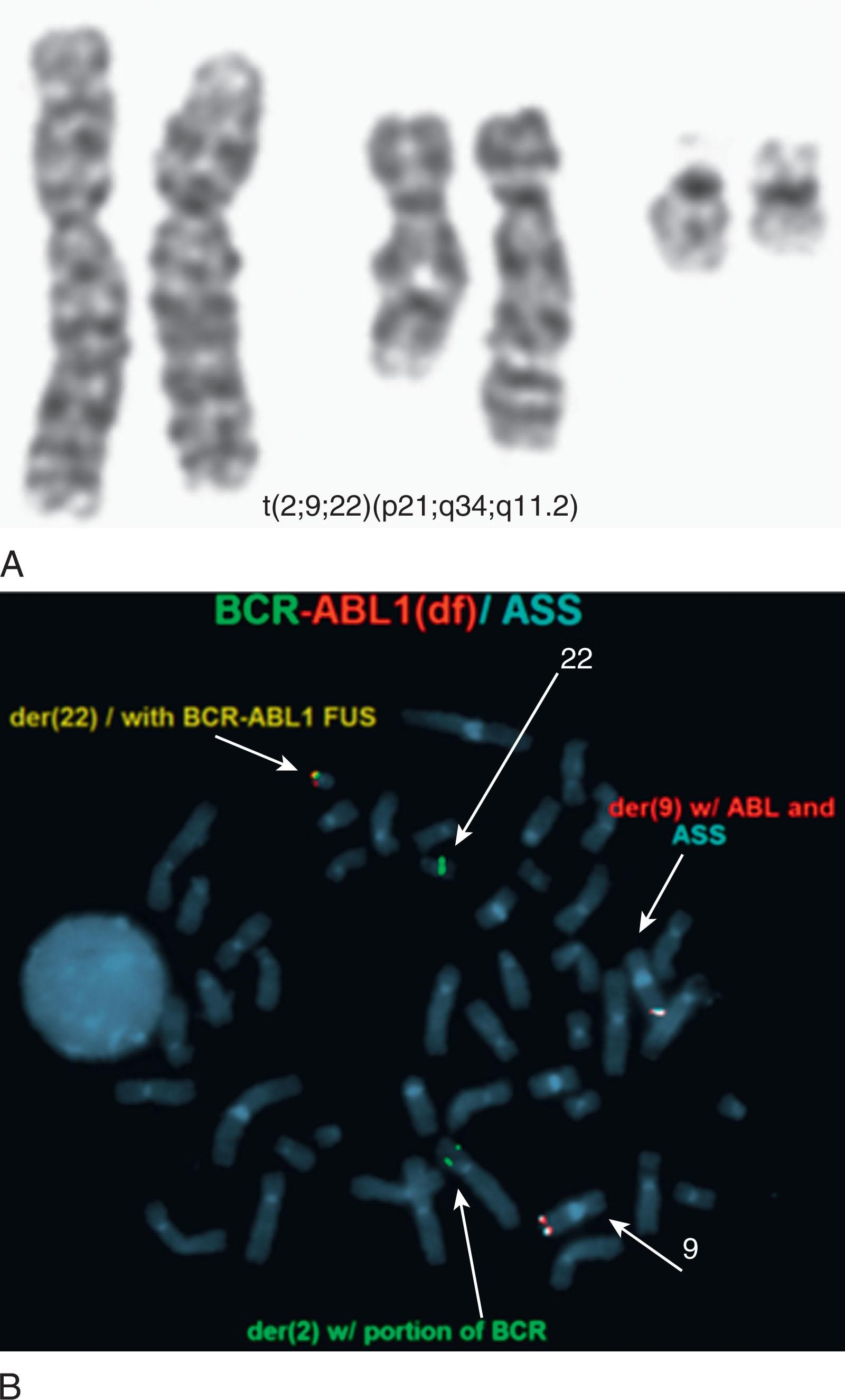
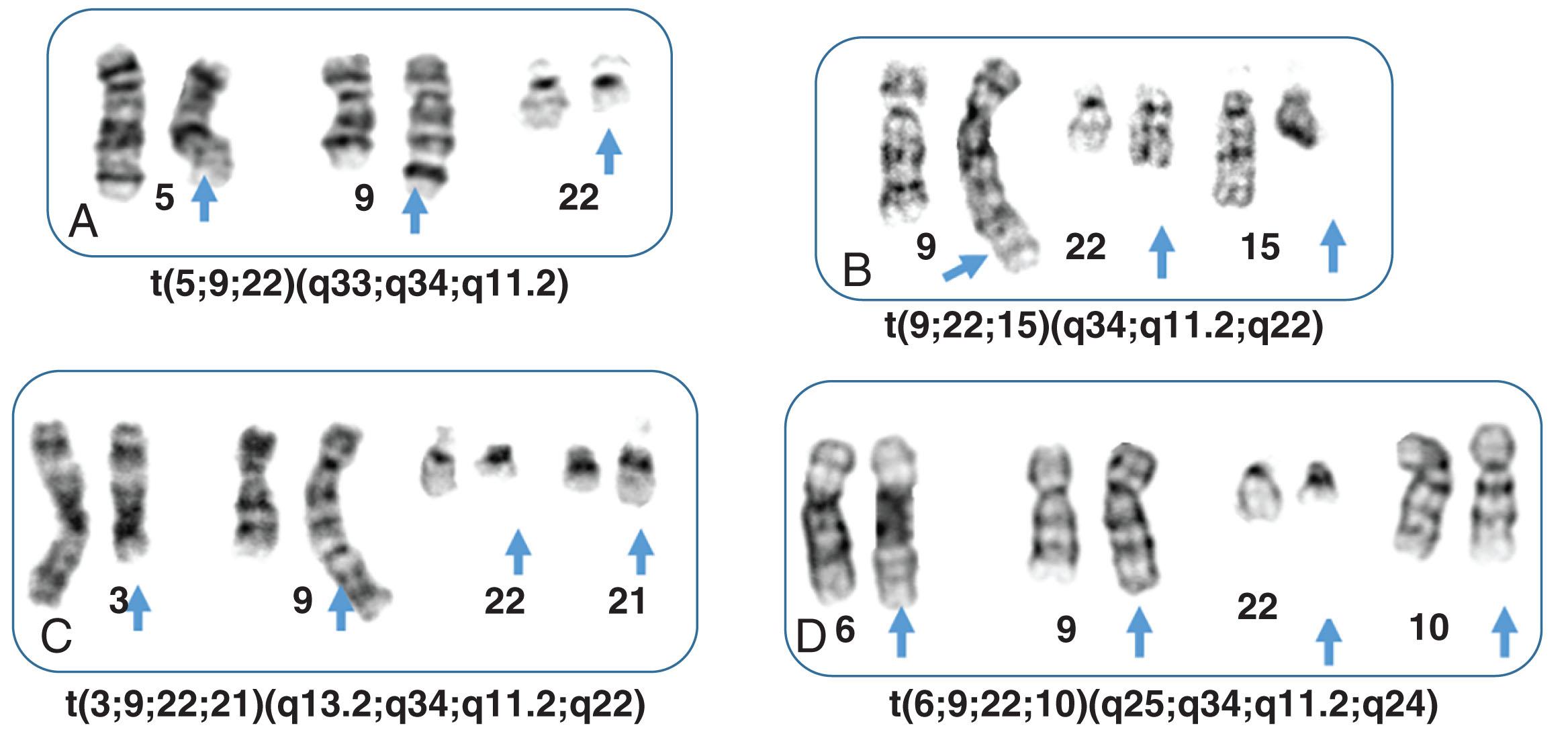
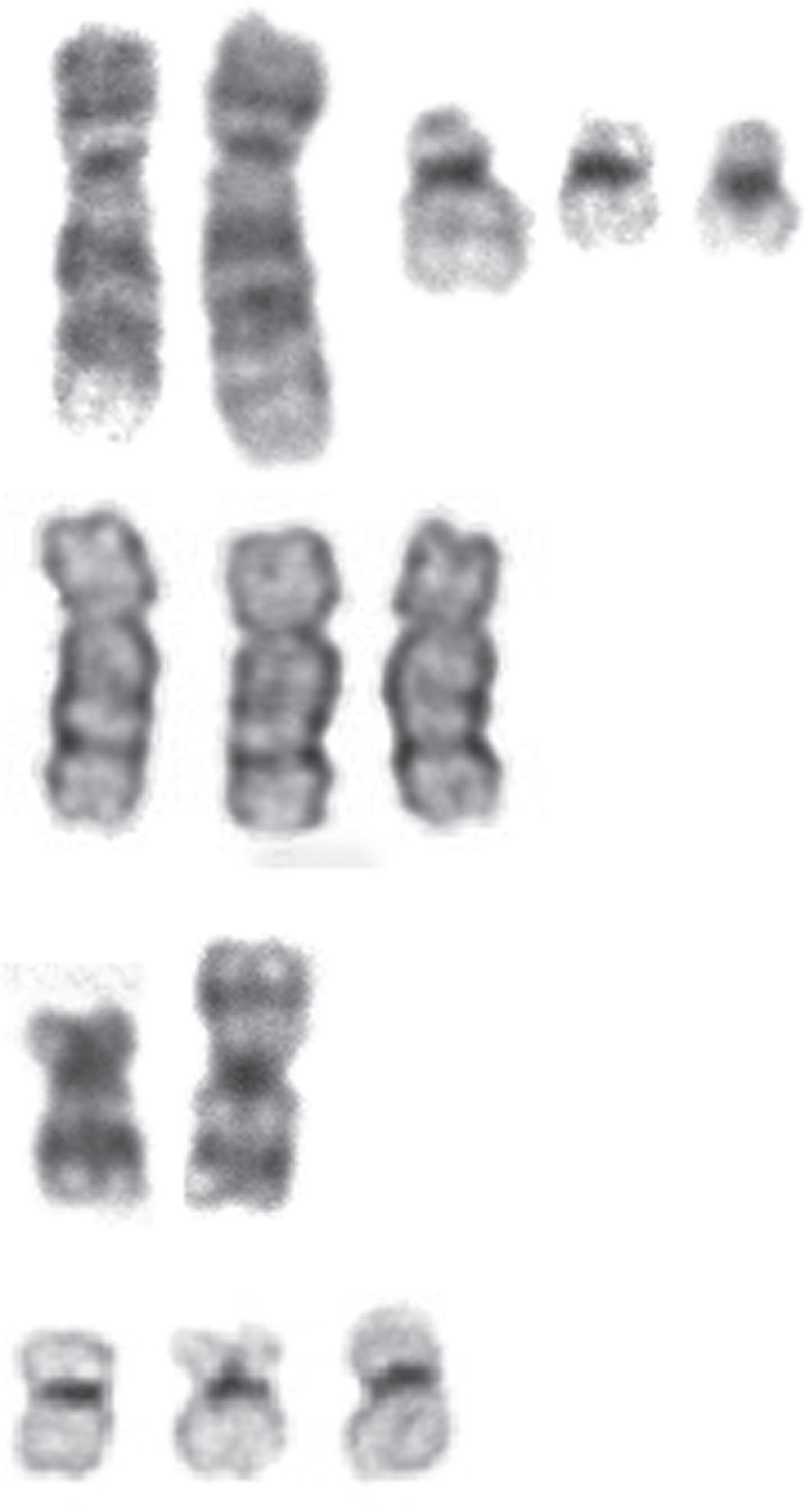
In 5% to 8% of newly diagnosed patients with CML, the Ph chromosome is present as a 3- or 4-way complex derivative Ph chromosome, always involving chromosomes 9 and 22 as well as an additional one or two chromosomes. In our own laboratory we have observed over many years at least 25 different complex Ph translocations. Three-way translocations are more frequent than 4-way translocations. Some of the examples are the following translocations ( Fig. 57.11 ): t(2;9;22)(p13;q34;q11.2), (9;22;7)((q34;q11.2;p15), t(9;19;22)(q34;p12;q11.2), t(5;9;22) (q35;q34;q11.2), t(9;22;8)(q34;q11.2;p21), t(9;11;22)(q34;p15;p11.2), t(9;22;22) (q34;q11.2;p11.2), t(6;9;22;10)(q25,q34;q11.2;q24), t(7;9;22;22)(q22;q34;q11.2;q13.2), t(6;9;22;11)(q25,q34;q11.2;q13). Most of these complex structural rearrangements are unique but some of them such as t(2;9;22) are recurrent. Moreover, there are less than 1% of CML patients in chronic phase who acquire the t(9;22) clone abnormalities as well as t(8;21), inv(16), t(11q23), t(15;17), and t(3;3) that are associated with AML. In one recent retrospective study among 1382 Ph-positive patients 0.36% of patients had co-occurrence of these abnormalities and most of them progressed to accelerated or myeloid blast crisis.
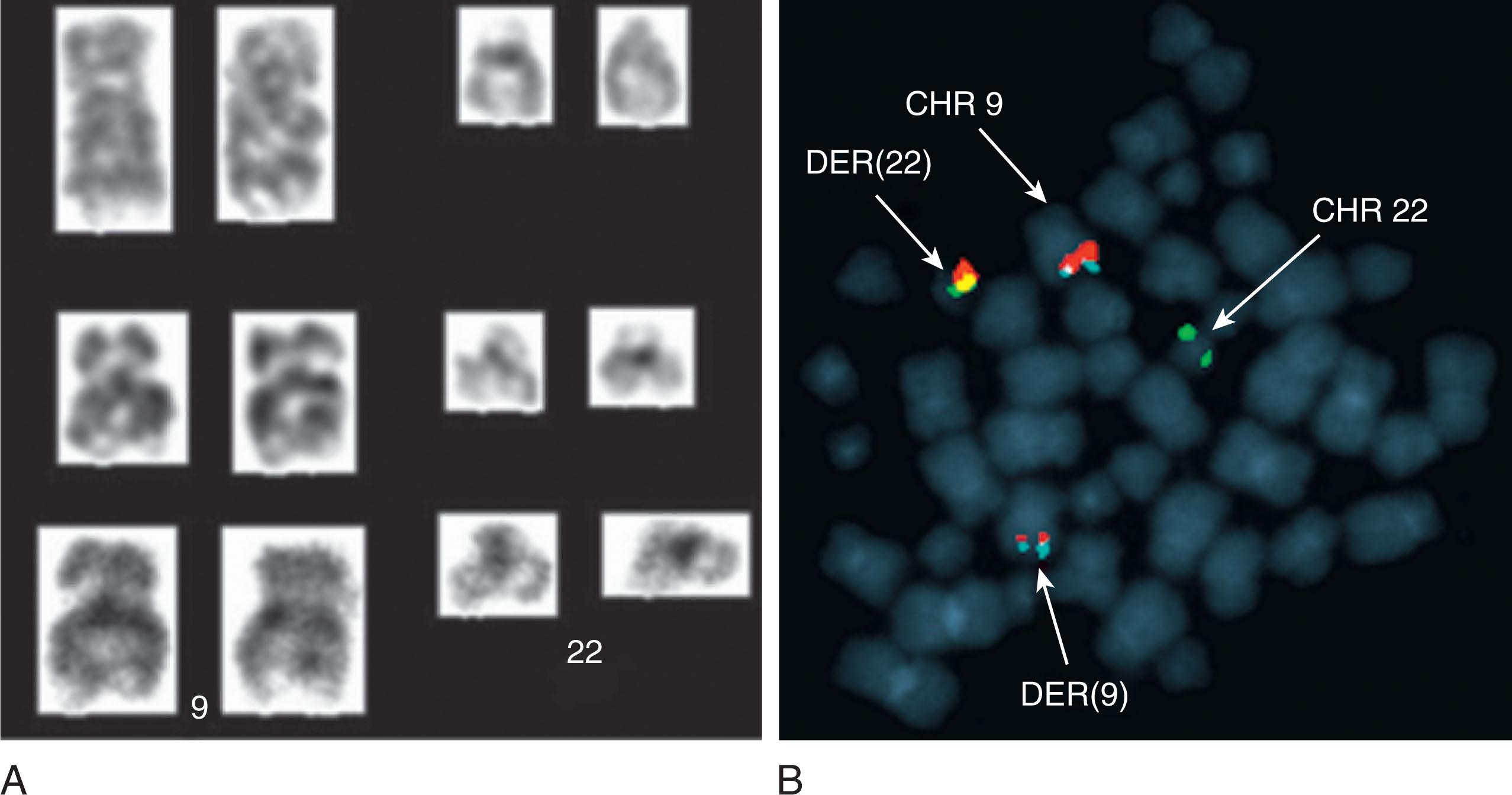
In about 3% to 5% of patients with CML who are Ph-negative by cytogenetic studies, the clonal and stem cell origin of these hematologic malignancies can still be demonstrated, and molecular analysis reveals the BCR-ABL1 fusion in approximately 1% ( Fig. 57.12 ). These patients have chromosomal insertions resulting in BCR-ABL1 fusion which can occur in multiple ways. Approximately 70% of cases exhibit BCR-ABL1 fusion located on a chromosome 22, which was either a cryptic der(22), or “cryptic Ph” or non-cryptic der(22) or “masked Ph.” The remaining cases have BCR-ABL1 fusion located on a cryptic or non-cryptic der(9). Of the 119 such cases reported in literature 77 patients (64.7%) had an insertion-derived BCR-ABL1 fusion as a cryptic or a masked Ph. This type of BCR-ABL1 fusion does not influence OS of these CML patients. In the majority of Ph-negative patients, an ABL1 insertion from chromosome 9 to 22q11.2 results in a BCR-ABL1 fusion product without reciprocal translocation of sequences from chromosome 22 to chromosome 9. Sizes of the inserts can vary, which may be closely related to the morphological appearance of the chromosomes with the BCR-ABL1 or those involved chromosomes without the BCR-ABL1 . Intensive FISH mapping using many probes targeting ABL1 , BCR , and their flanking regions determined that a classic t(9;22) usually involves an exchange of approximately 10 MB of 9q (from the ABL1 to the 9q telomere) and ∼25 MB of 22q (from BCR to the 22q telomere).
Approximately 2% of patients truly are Ph-negative and BCR-ABL fusion-negative. These patients may not have CML but rather another MPN ( Chapter 71 ). The concept that the BCR-ABL1 fusion plays a central role in the pathogenesis of CML is strongly supported by two lines of evidence: (a) retroviral transduction experiments in which P210 BCR-ABL1 is expressed in murine marrow cells, resulting in an MPN resembling CML, and (b) the fact that imatinib, a TKI, selectively inhibits the BCR-ABL1 fusion protein in mice and specifically inhibits the growth of human Ph-positive cells in vitro and in vivo. Although considered necessary, BCR-ABL1 may not be the initial event or be sufficient to cause the malignant transformation resulting in CML (see earlier section on clonal origin).
PCR analysis can determine the exact breakpoints of DNA fusion products. Reverse transcriptase PCR (RT-PCR) and Northern blot analysis allow detection of BCR-ABL1 transcripts at the RNA level. Current FISH studies can detect BCR-ABL1 fusion at diagnosis using a dual-color BCR-ABL1 ES probe or BCR-ABL1 dual-fusion probe (see Fig. 57.12 , see also Fig. 57.9 ). A triple-color BCR-ABL1-ASS probe is used for detection of deletions on both chromosomes 9 and 22 ( Fig. 57.13 ). Approximately 12% to 15% of patients with CML have large deletions adjacent to the Ph chromosome translocation breakpoint on the derivative 9 chromosome. These deletions are heterogeneous and may involve both chromosomes 9 and 22 (majority of cases), only chromosome 9 (8% of patients with deletions), or only chromosome 22 (4% of cases with deletions). Moreover, deletion size is variable, ranging from 0.5 Mb to greater than 10 Mb. A more refined study applying SNP microarrays revealed three common deleted regions: (1) a 162-kb loss at 9q34, (2) a 138-kb deletion at 22q11.2, and (3) a 102-kb deletion at 22q11.2. It appears that the partial deletion of the ABL1-BCR fusion on derivative 9 chromosome occurs as a part of the same process as the formation of the BCR-ABL1 translocation. Before imatinib therapy, these deletions were associated with an adverse prognosis; however, in a study of 521 patients with CML, the cumulative incidence of complete cytogenetic responses and major molecular responses and the overall survival (OS) were comparable after 5 years of follow-up between CML patients with and without the del(9q).
At diagnosis, conventional cytogenetics remains the gold standard because the chromosome analysis will identify not only the t(9;22) but also other chromosomal abnormalities that may indicate accelerated or blast phase of the disease or clonal proliferation of Ph-negative cells.
Imatinib mesylate (Gleevec) has revolutionized therapy for CML and, for most patients, has transformed a deadly disease into a chronic disorder that is compatible with normal life. The standard method for monitoring a patient’s response to therapy is conventional cytogenetic analysis of cells obtained from a marrow aspirate. In the phase III international Randomized and STI-571 (IRIS) study, 89% of patients had Ph-negative marrow aspirates as determined by conventional cytogenetics after 5 years of treatment. However, conventional cytogenetics has limited sensitivity. The degree of MRD is determined to be an important prognostic factor for patients with CML on therapy. Patients with CML without a discernible Ph chromosome detected by conventional cytogenetics analysis may still harbor up to 10 10 leukemic cells.
Interphase FISH does not depend on the cycling status of cells, and use of double-fusion probes has reduced false-positive results to approximately 1%. However, if peripheral blood cells rather than marrow aspirate cells are used to monitor MRD, the high percentage of BCR-ABL1 fusion-negative lymphoid cells may underrepresent the actual residual tumor load. In most direct comparison studies, interphase FISH of peripheral blood compared with conventional cytogenetics of marrow in patients who are treated with imatinib showed good correlations ( r = 0.91 to 0.97). Real-time quantitative PCR (RQ-PCR) is by far the most sensitive method for detecting MRD. It provides an accurate measure of the total leukemia cell mass and the degree to which BCR-ABL1 transcripts are reduced by therapy, and it correlates with progression-free survival.
The goal of therapy in CML is to achieve a molecular remission as measured by the reduction or elimination of BCR-ABL1 transcripts. In the IRIS study, at 5-year follow-up, complete cytogenetic response combined with major molecular response at 12 months was associated with a 97% progression-free survival rate. This compares with an 89% progression-free survival for those with complete cytogenetic response but without a major molecular response. Current international recommendations for optimal molecular monitoring of patients receiving imatinib treatment includes an RQ-PCR assay expressing the BCR-ABL1 transcript levels on an internationally agreed upon scale. The term major molecular response corresponds to ≤0.0% BCR-ABL1, whereas the designation complete molecular response should be used only for patients with undetectable BCR-ABL1 , where the limit of detection is confirmed to be at least a 4.5 log reduction from baseline levels. The two major obstacles to successful imatinib-based therapy for patients with Ph-positive, BCR-ABL1 fusion-positive CML are the persistence of BCR-ABL1 fusion-positive cells and relapse of the disease because of emergence of resistance to imatinib. Acquired resistance to imatinib treatment can be due to two events: amplification of BCR-ABL1 fusion product ( Fig. 57.14 ) and mutations in the ABL kinase domain. Currently, 100 different ABL1 kinase domain mutations have been described, although only about 15 are common and account for more than 85% of all mutations detected.
Patients with CML who cannot tolerate or are resistant to imatinib may benefit from the second and now third generation of TKIs ( Chapter 69 ) such as dasatinib, nilotinib, bosutinib, and ponatinib. These agents bind to the ABL kinase domain in a matter distinct from that of imatinib and thereby retain activity against nearly all imatinib-resistant mutations. Recent meta-analysis revealed ABL mutation rates with imatinib 9.7%, dasatinib 1.7%, nilotinib 3.3%. The most common specific mutations were T315I, E255k, and M315I. T315I mutations constitute 58% of dasatinib-related mutations and 13% of imatinib-related mutations. These mutations inhibit the binding of the TKIs, hindering the treatment of patients with CML. The introduction of ponatinib, a new formulation of TKI, has been shown to overcome resistance incurred by the T315I mutation (see Chapter 69 ).
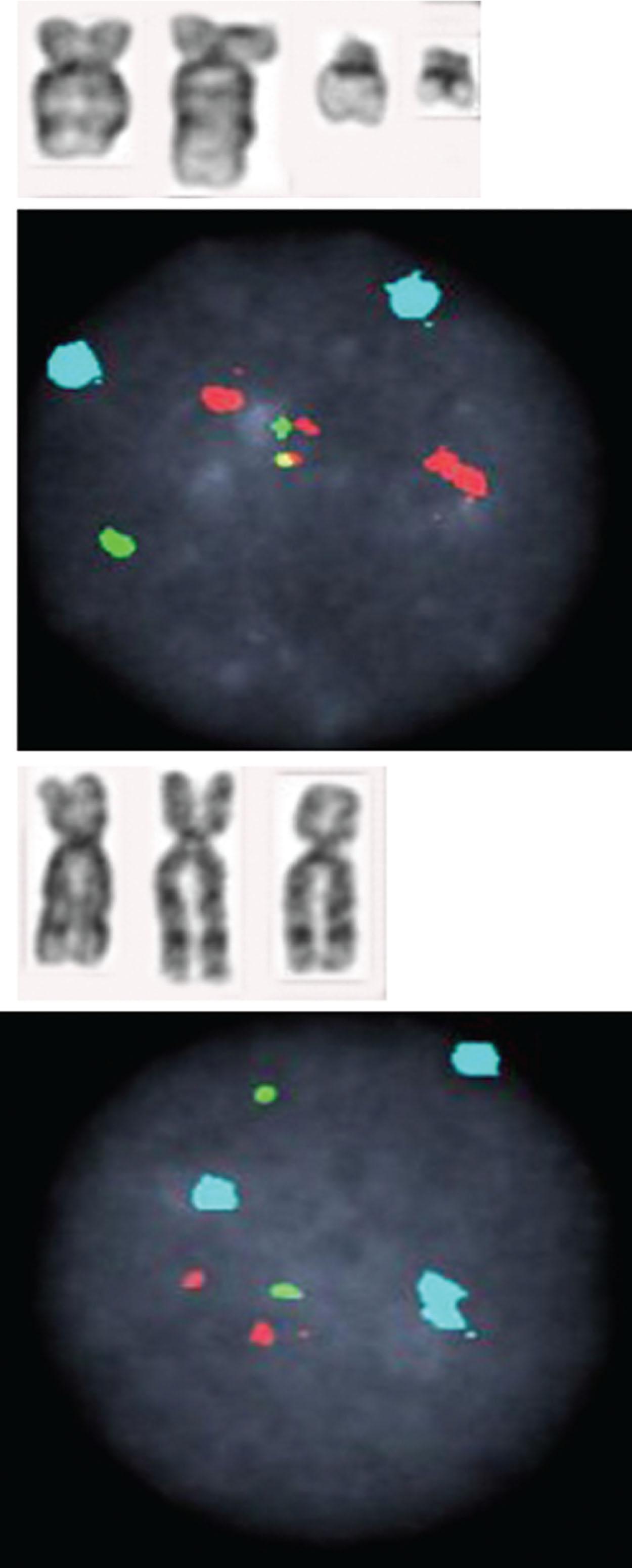
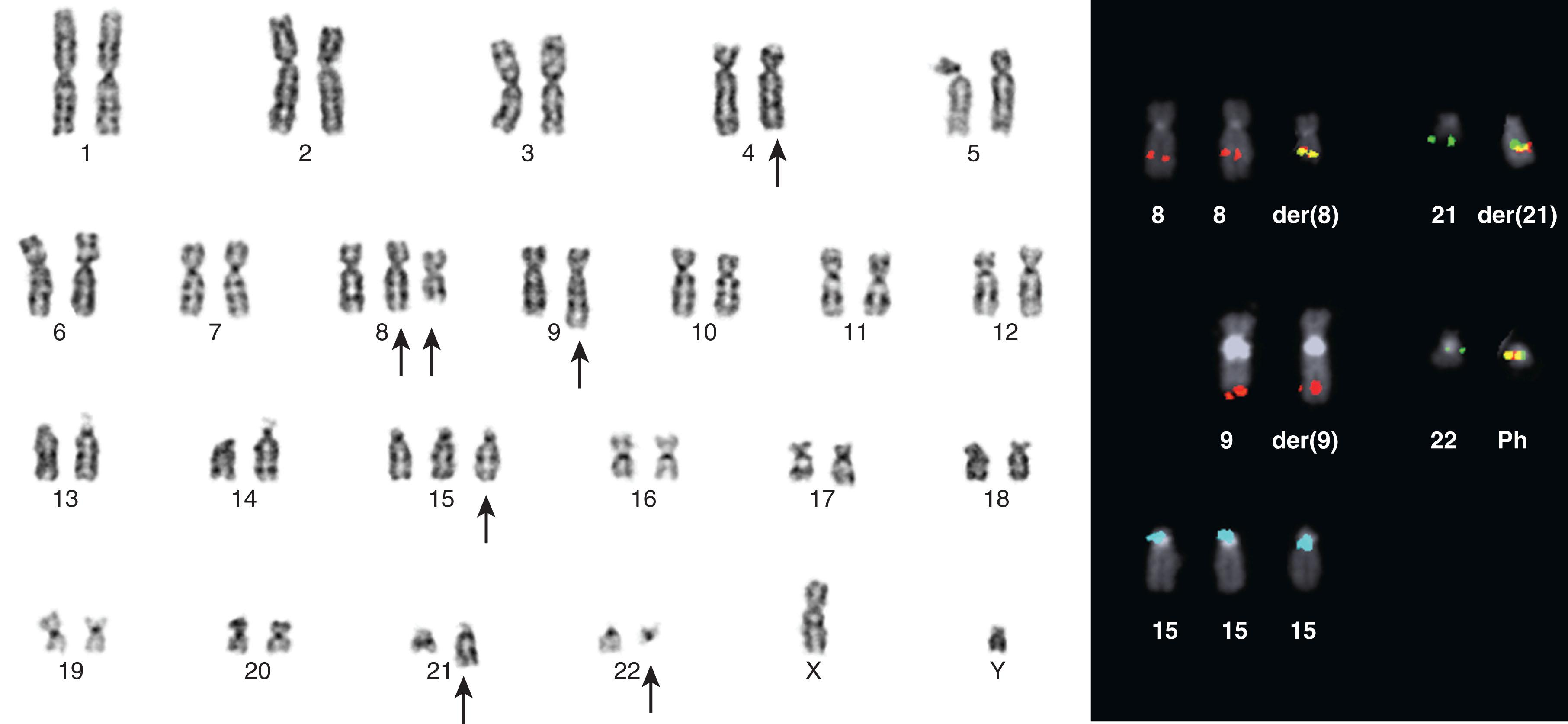
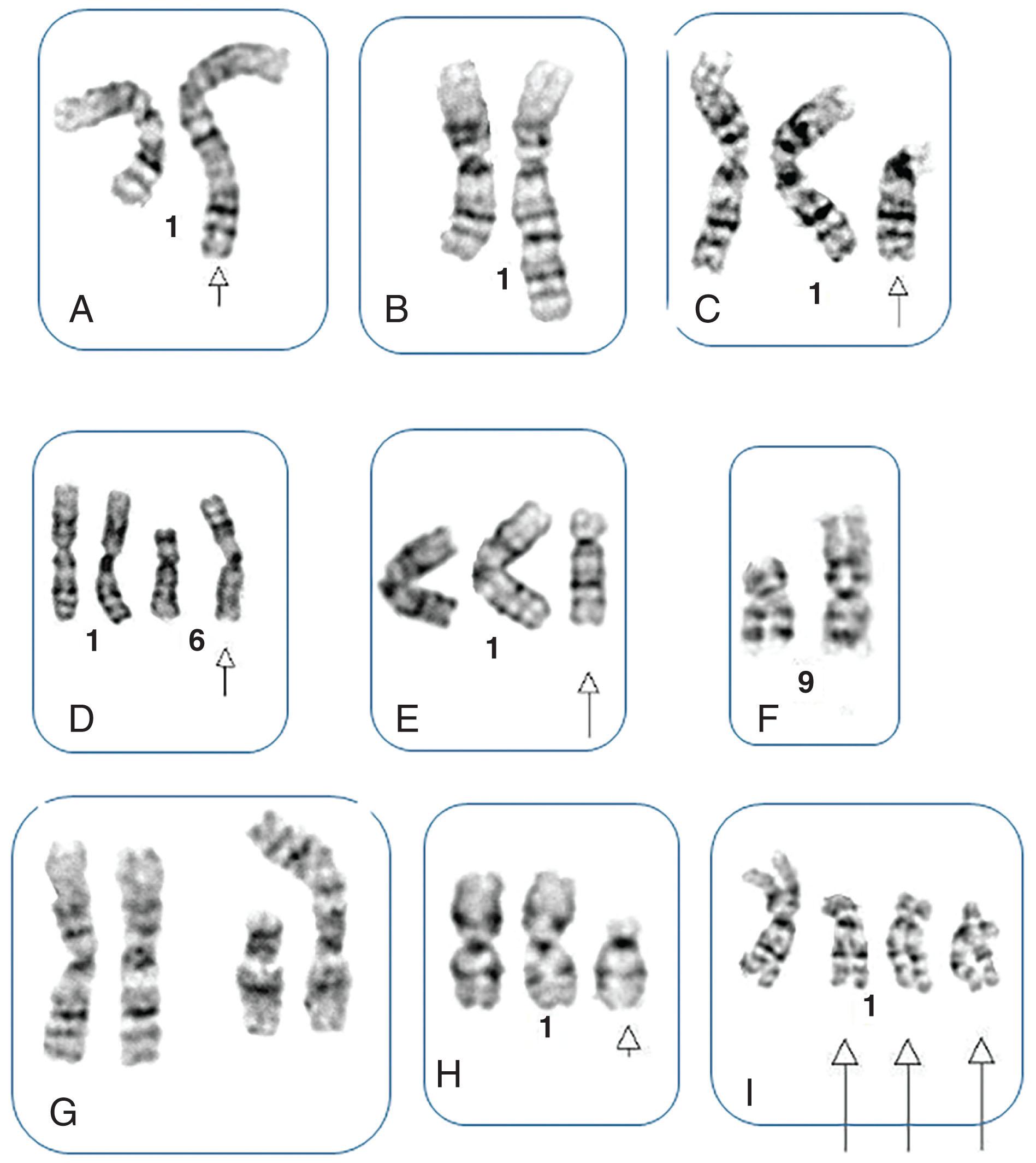
Between 5% and 8% of patients undergoing treatment with imatinib will develop additional chromosomal abnormalities (ACAs) such as trisomy 8, monosomy 7, del(20q), and other anomalies in BCR-ABL1 fusion-negative cells. Imatinib may induce chromosomal abnormalities in BCR- ABL1 − cells. Alternatively, imatinib may uncover chromosomal abnormalities present before therapy after significant reduction of Ph-positive cells has occurred ( Fig. 57.15 ). Presence of +8 and other chromosomal anomalies in Ph-negative cells in patients treated with imatinib suggests that CML has a multistep pathogenesis and that clonal Ph-negative cells precede the development of the Ph-positive clone ( Fig. 57.16 ). This important observation about the pathogenesis of CML demonstrates the power of conventional cytogenetics, even in the era of molecular assays, and should be used at least annually while patients are undergoing TKI treatment. This hypothesis that clonal development occurs in Ph-negative cells before development of the Ph chromosome was recently confirmed using NGS by targeting 25 genes frequently mutated in myeloid disorders. Ph-negative clones were analyzed in 14 patients who developed clonal cytogenetic abnormalities during TKI treatment. Mutations affecting DNMT3A , EZH2 , RUNX1 , TET2 , TP53 , U2AF1 , and ZRSR2 were detected in 43% of these patients. In two patients, the mutations were also found in corresponding Ph-positive diagnostic samples. Moreover, somatic mutations in addition to BCR-ABL1 affecting ASXL1 , DNMT3A , RUNX1 , and TET2 genes are found in 33% of patients. When individual hematopoietic colonies were analyzed from patients with CML at diagnosis, most mutations were present in the Ph-positive cells. By contrast, NGS of subsequent samples during TKI treatment revealed that one DNMT3A mutation occurred in Ph-negative cells that was also present in Ph-positive cells at diagnosis, strongly implying that DNMT3A preceded the appearance of the BCR-ABL1 rearrangement which provides support for the initial hypothesis that BCR-ABL1 and the formation of the Ph chromosome were not the initiating events in the pathogenesis of CML. Another example of clonal evolution in MPNs evolving over time is the co-occurrence of the JAK2V617F mutation and BCR-ABL1 . These rare patients usually have a long (10 to 20 years) history of JAK2V617F+ polycythemia vera (PV) before BCR-ABL1 and the Ph chromosome is acquired. Single cell genotyping revealed that both JAK2V617F and BCR-ABL1 can occur concurrently in hematopoietic stem cells and that JAK2V617F+ occurs before the acquisition of BCR-ABL1 . The contribution of BCR-ABL1 to disease progression appears to be greater than that of JAK2V617F , since these patients display a clinical phenotype that is consistent with CML rather than PV. Recent studies have demonstrated the presence of somatic mutations in TET2 , DMNT3A , and ASXL1 in both diagnostic and/or follow up remission CML specimens implying evolution within the Ph-negative clone and indicating that these mutations are early events which preceded formation of the BCR-ABL1 gene. The mechanism of transformation to advanced-phase CML is complex and varied and remains poorly understood. In myeloid blast crisis (70%) of CML patients show karyotypic evolution, that is, new chromosomal abnormalities in very distinct patterns in addition to the Ph chromosome. The most common abnormalities include gain of chromosome 8 (33%) or +19 (12%), gain of a second Ph chromosome (30%), i(17q)(20%), alone or in combination, to produce modal chromosome numbers of 47 to 50 ( Figs. 57.17 and 57.18 ). In males, LOY is frequently observed. Isochromosome 17q occurs almost exclusively in myeloid blast crisis. Other less frequently observed abnormalities include monosomy of chromosomes 7 and 17, and trisomy of chromosomes 17 and 21. In addition to these common karyotypic evolutions, additional chromosomal aberrations specific for AML—for example, t(8;21) ( Fig. 57.19 ), inv(16), t(3;21)(q26;q22), and most frequently, inv(3q)/t(3;3), resulting in the overexpression of MECOM (EVI1) —have been observed. Simultaneous presence of t(9;22) and inv(3) at diagnosis or following TKI therapy appears to be associated with a lack of response to treatment and a clinical course is similar to that of cases with inv(3) without any other chromosomal abnormality. In patients treated with TKI, these ACAs have been classified into two groups based on their impact on survival. ACAs in the Good prognostic group include trisomy 8, LOY, and an extra copy of the Ph chromosome. The second group includes chromosomal abnormalities that are associated with a relatively poor prognosis including i(17q), −7/del(7q), and 3q26.2 rearrangements. The concurrent presence of two or more ACAs was associated with an inferior survival. A prospective study of 1551 imatinib-treated chronic phase patients (German CML-study IV) defined more precisely a high-risk and a low-risk group based on the presence of ACAs. High-risk chromosome abnormalities and their impact on survival included +9,+Ph, i(17q),+17,+19, 3q26.2, 11q23, −7/del(7q), and a complex karyotype, while the low-risk group included all other karyotypic changes. High-risk abnormalities were observed in 6% of patient during chronic phase but in 61% of patients who progressed to blast crisis while low-risk chromosome abnormalities were observed in 21% during CP and in 2.8% of patients who progressed. Most importantly, this study demonstrated that the combination of high-risk cytogenetic abnormalities and a low-level blast cell count (1% to 5%) was associated with disease progression and death from CML. Therefore, additional high-risk chromosomal abnormalities are an early marker of CML progression even when present in patients with low numbers of blast cells.
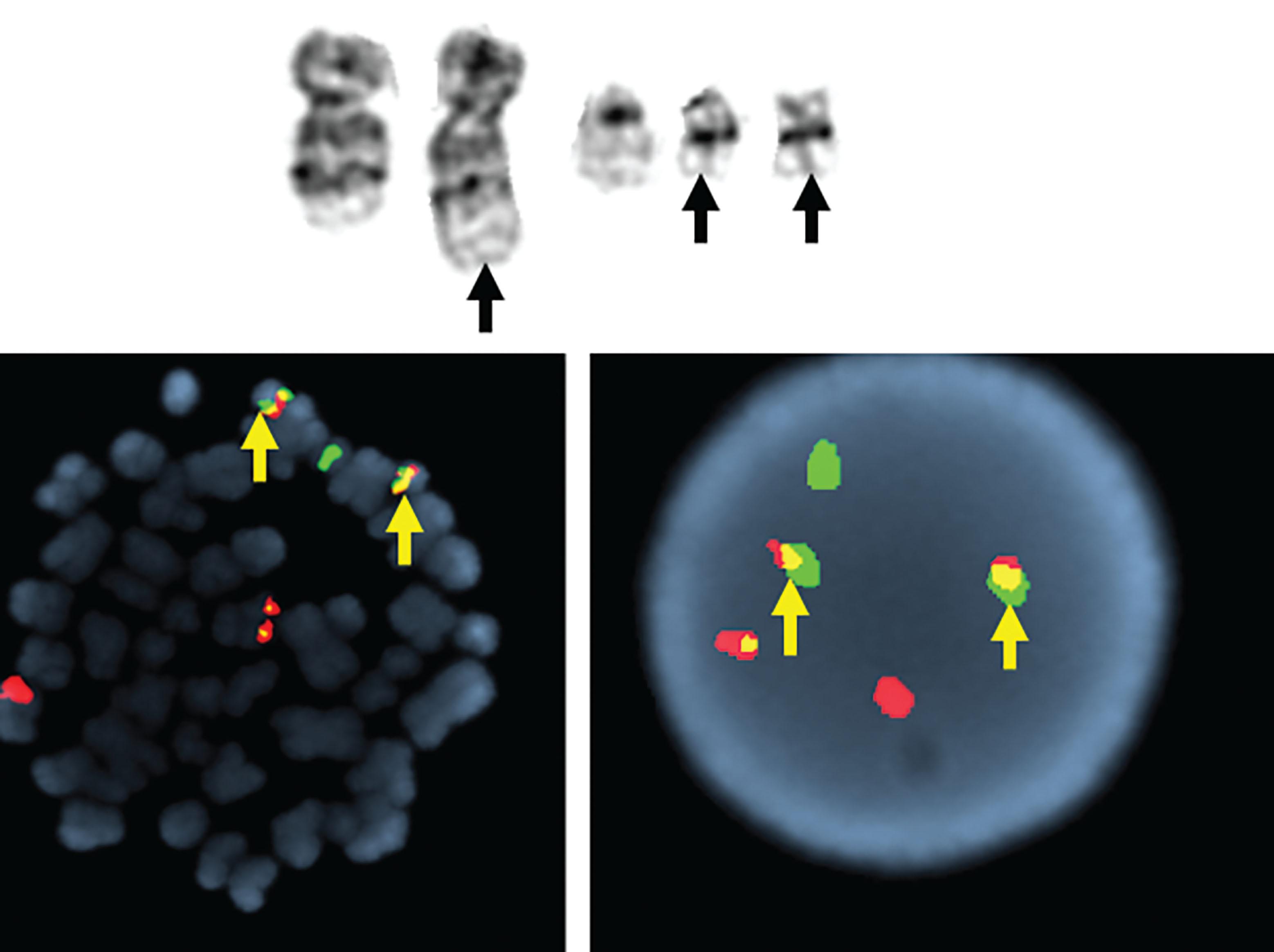
Complete cytogenetic remission for patients in accelerated phase or blast crisis CML treated with imatinib is rarely accompanied by a normal karyotype; however, 6% to 17% of these patients may have some cytogenetic response (see box on Genetic Testing for Chronic Myeloid Leukemia ).
High-resolution cytogenomic studies have demonstrated that in contrast to the chronic phase CML (which is characterized only by genomic imbalances in or around BCR and ABL1 , specifically deletions), during the blast phase, many additional genomic imbalances occur. Using aCGH, 44 patients in chronic phase CML as well as 11 in myeloid and in lymphoid blast crisis were investigated, and a spectrum of recurrent genomic imbalances was associated with disease progression, including losses at 1p36, 5q21, 9p21, and 9q34 and gains at 1q, 8q24, 9q34, 16p, and 22q11. The most common mutations in myeloid blast phase involve tumor suppressor gene TP53 (about 25% of cases) and the RUNX1 in about 40% of cases. Moreover, the MYC oncogene has been recently linked to BCR-ABL1 activity and CML progression to myeloid blast crisis. Recently, mutations in genes frequently found in Ph-negative MPNs such as CBL , TET2 , ASXL11 , and IDH1/2 were also identified in patients in accelerated/blast phase. Additionally, deletions of RB1 , gain of function mutations in GATA-2 and RAS are just some of the spectrum of cytogenomic changes characterizing progression of CML.
At diagnosis of CML, perform quantitative cytogenetic analysis using the bone marrow aspirate, which is the sine qua non because peripheral blood cells rarely contain sufficient numbers of mitotic cells at the time of presentation. If the bone marrow aspirate is a “dry tap,” perform interphase FISH using a BCR-ABL1 extra-sensitive dual-fusion color probe. According to the ELN 2020 and British Society of Haematology 2020 recommendations, a full bone marrow karyotype is important also to determine the phase of the disease. Their recommendations also include mandatory molecular work to identify the BCR-ABL1 fusion transcript type for molecular monitoring to guide future management. To monitor patients with CML during therapy, use FISH of blood or bone marrow to track changes in the percentage of cells with BCR-ABL1 fusion at 3-month intervals. If the molecular assay demonstrates BCR-ABL1, but the Ph cannot be identified by cytogenetics, a FISH test is required. Once the patient is in complete cytogenetic and FISH remission, the consensus recommendation is to perform real-time quantitative polymerase chain reaction and to follow the patient at 3-month intervals until molecular remission is achieved. According to ELN 2020 recommendations, greater than 10% BCR-ABL1 at 3 months indicates treatment failure. BCR-ABL1 transcript level at less than 0.1% is defined as major molecular response remission. Early indicators of progression are the appearance of additional chromosomal abnormalities and somatic mutations. At relapse, perform a cytogenetic analysis to assess the karyotype of the malignant clone and to determine if a new chromosomally abnormal clone has developed or a new subclone has evolved from the Philadelphia chromosome–positive clone.
Patients with CML may present initially in lymphoid blast crisis mimicking Ph-positive B-cell ALL. Although both the CML and Ph-positive ALL (see later) are driven by the constitutively active BCR-ABL1 tyrosine kinase, their prognoses differ significantly and it is important to differentiate these two entities. Cytogenomic analysis of 78 CML patients in lymphoid blast crisis demonstrated a unique signature of genomic deletions within the Ig heavy-chain and TCR genes, indicating that their presence is essential for the development of a malignant clone with a lymphoid phenotype. Moreover, 50% of patients in lymphoid blast crisis had mutations in cyclin-dependent kinase inhibitor CDKN2A , 25% to 35% in RUNX1 , and ∼15% to 25% have mutation in the BCOR gene, which interacts with histone deacetylates, a known apoptosis regulator. One of the most important differences between lymphoid and myeloid blast crisis is the exclusive interaction with STAT proteins. STAT1 is activated mainly by p190, while STAT3 and STAT5 are preferentially activated by the P210 isoform.
The precise causes and mechanisms responsible for CML progression are still elusive and current thinking is that progression of CML is the result of increased and complex genomic instability combined with heterogeneity of numerous coexisting mutations and signaling disruptions causing defective or an insufficient ability to repair DNA.
Ph-negative MPNs are disorders arising in a single clone of multipotent precursor cells in which one or all myeloid lineages are abnormally amplified. Classic and more frequently encountered Ph-negative MPNs include PV, primary myelofibrosis, and essential thrombocytopenia (ET). Their cytogenetic and genomic profiles are described in Chapters 70–73 . However, included in this chapter are Fig. 57.20A and B and 57.21 in order to illustrate the most frequent abnormalities associated with the Ph-negative MPN. Different 9p chromosomal abnormalities result in JAK2 numerical gain (see Fig. 57.20 ) while nine examples of 1q gain, associated with disease progression, are shown in Fig. 57.21 .
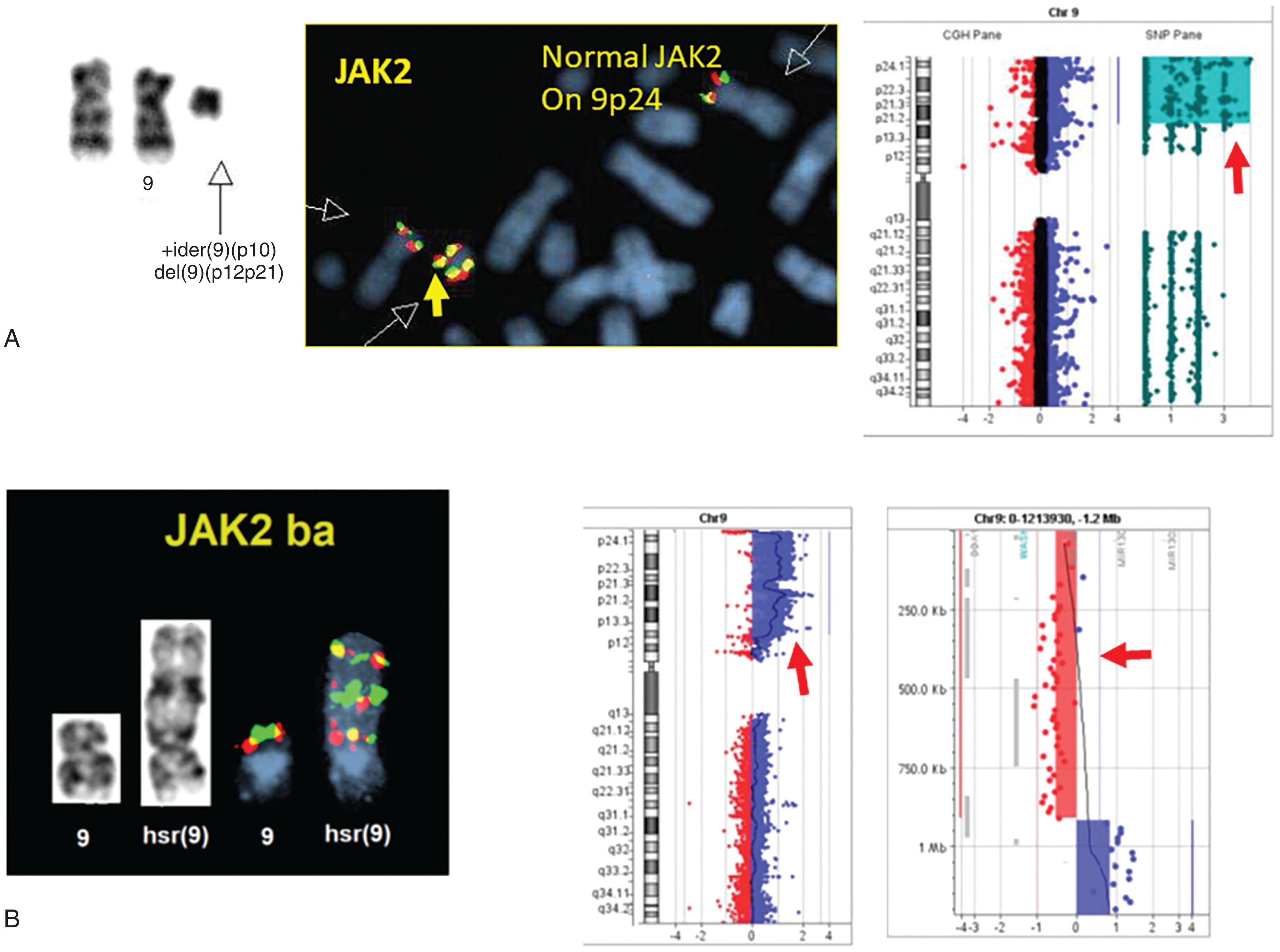
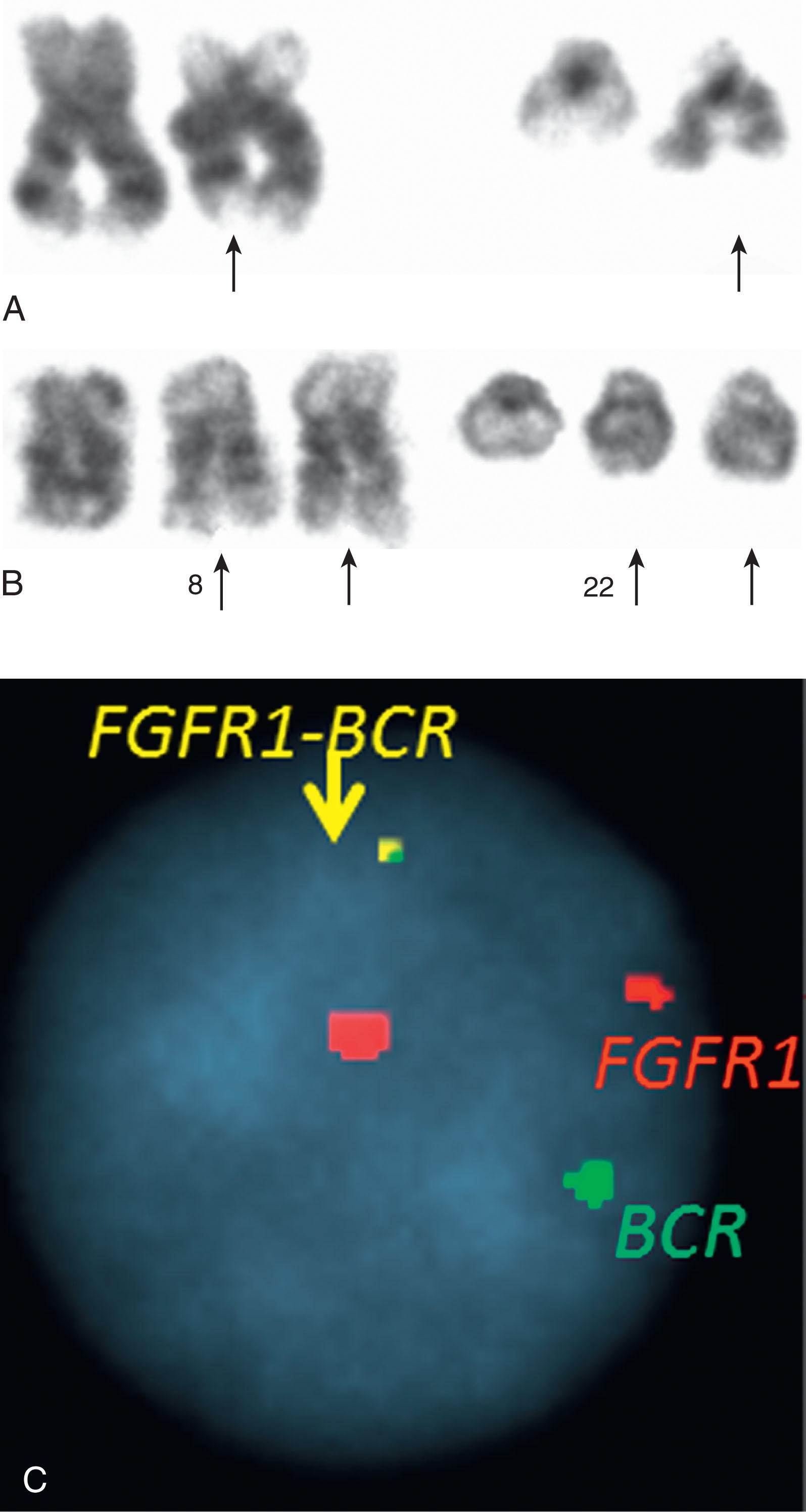
Less frequently encountered similar disorders include chronic neutrophilic leukemia ( Chapter 72 ), hypereosinophilic syndrome/chronic eosinophilic leukemia ( Chapter 74 ), systemic mastocytosis ( Chapter 75 ), atypical CML, and unclassifiable MPNs ( Chapter 72, Chapter 73, Chapter 74, Chapter 75 ). Rare recurrent balanced abnormalities in atypical Ph-negative MPNs can involve the PDGFRB gene localized at 5q33, FGFR1 gene on 8p11, and PDGFRA gene on 4q12. The best described are t(5;12)(q33;p13), resulting in ETV6-PDGFRB fusion protein, and t(8;13)(p11;q12), resulting in the ZNF198-FGFR1 fusion gene. The disorder known as 8p11 myeloproliferative syndrome has been classified by WHO as belonging to the group of myeloid/lymphoid neoplasms associated with eosinophilia and gene rearrangements. Some of these diseases are related to fusion genes between FGFR1 located on 8p11 and various translocations fusing at least 14 different 5′ partner genes to the 3′ part of the FGFR1 gene that encodes tyrosine kinase domain ( Table 57.4 ). They include TPR (1q25), LRRFIP1 (2q37), SQSTM1 (5q35) FGFR10P (6q27), TRIM24 (7q34), CUX1 (7q22), PLAG1 (8q12), CEP110 (9q33), NUP98 (11p15 ), FGFR1OP2 (12p11), CPSF6 (12q15), ZMYM2 (13q12), MYO18A (17q23), HERVK (19q13), and BCR (22q11) ( Fig. 57.22 ). The most frequent translocation is t(8;13), and cases with the submicroscopic deletion of the 5′ part of the FGFR1 gene have also been reported, similar to the formation of t(9;22) with deleted sequences from der(9q) and/or 22q identified in CML. The presence of 8p11 abnormalities in both myeloid and lymphoid cells suggests its origin from a common stem cell.
| Chromosomal Abnormality | Genes Involved | MPD Entity |
|---|---|---|
| t(9;12)(q34;p13) | ETV6-ABL | CML-like, T-ALL |
| 5q32 | PDGFRB | Associated translocations |
| t(1;5)(q25;q32) | TPM3-PDGFRB | Atypical MPN |
| t(2;5)(p21;q32) | SPTBN1-PDGFRB | Atypical MPN |
| t(4;5)(q21;q32) | PRKG2-PDGFRB | Atypical MPN |
| t(5;7)(q32;q11) | HIP1-PDGFRB | CMML-like |
| t(5;10)(q32;q21) | CCDC6-PDGFRB | Ph-negative MPN |
| t(5;12)(q32;p13) | ETV6-PDGFRB | CEL, CMML |
| t(5;12)(q32;p13.3) | ERC1-PDGFRB | Atypical MPN |
| t(5;14)(q33;q24) | NIN-PDGFRB | Atypical MPN |
| t(5;14)(q32;q32) | KIA1509-PDGFRB | Atypical MPN |
| t(5;14)(q32;q32) | TRIP11-PDGFRB | AML |
| t(5;15)(q23;q15) | TP53BP1-PDGFRB | CEL |
| t(5;15)(q32;q22) | TPS3BP1-PDGFRB | Atypical MPN |
| t(5;16)(q32;p13) | NDE1-PDGFRB | Atypical MPN |
| t(5;17)(q32;p11) | HCMOGT1-PDGFRB | Juvenile CMML |
| t(5;17)(q32;p11,2) | MYO18A-PDGFRB | Atypical MPN |
| t(5;17)(q32;p13) | RABEP1-PDGFRB | CMML |
| t(5;17)(q32;q21) | COL1A1-PDGFRB | Atypical MPN |
| 8p11 | FGFR1 | Associated translocations |
| t(2;8)(q37;8p11) | LRRFIP1-FGFR1 | MPN syndrome |
| t(6;8)(q27;p11) | FGFR1OP-FGFR1 | Stem cell MPD |
| t(7;8)(q32;p11) | TRIM24-FGFR1 | MPN |
| t(7;8)(q22;p11) | CUX1-FGFR1 | MPN syndrome |
| t(8;9)(p11;q22) | CEP110-FGFR1 | MPN syndrome |
| t(8;11)(p11;p15) | Nup98-FGFR1 | MPN syndrome |
| t(8;12)(p11;p11) | FGFR10P-FGFR1 | MPN syndrome |
| t(8;12)(p11;q12) | CPSF6-FGFR1 | MPN syndrome |
| t(8;13)(p11;q12) | ZMYM2-FGFR1 | MPN syndrome, leukemia, lymphoma |
| t(8;17)(p11;q11) | MYO18A-FGFR1 | MPN syndrome |
| t(8;19)(p11;q13) | HERVK-FGFR1 | MPN syndrome |
| t(8;22)(p11;q11.2) | BCR-FGFR1 | Lymphoproliferative disorder |
| 4q12 | PDGFRA | |
| del(4)(q12;q12) | FIP1L1-PDGFRA | CEL |
| t(4;22)(q12;q11.2) | BCR-FGFR1 | Atypical CML |
| 4q12 | KIT | SM |
| 9p24 | JAK2 | |
| t(9;22)(p24;q11.2) | BCR-JAK2 | CML-like |
| t(9;12)(p24;p13) | JAK2-ETV6 | CML-like |
| der(9)t(9;12)(p24;q13) | JAK-NF-E2 | MDS |
| t(8;9)(p22;p24) | PCM1-JAK2 | CMPD, AL |
| der(9;18)t(p13;p11) | Not reported | PV, PV→MF |
| der(9;18)(p10;q10) | Not reported | PV, PV→MF, ET→AML |
| der(9)t(1;9)(q12;q12) | Not reported | PV, MF |
| der(1;7)(q10;p10) | Not reported | ET |
| t(12)(q21 or q21) | Not reported | MF |
In patients with chronic eosinophilic leukemia/hypereosinophilic syndrome ( Chapter 74 ) the most frequent recurrent abnormality is the cryptic deletion on 4q12 as a result of FIP1L1-PDGFRA fusion gene reported to occur in from 3% to 56% of cases. The disparity in frequencies reflects differing levels of stringency criteria in the diagnosis of disorders with hypereosinophilia as well as different technologies used for detection of FIP1L1-PDGFRA fusion. An investigation of 376 patients with persistent unexplained hypereosinophilia revealed an 11% incidence of the FIP1L1-PDGFRA fusion gene detected using highly sensitive RQ-PCR. Patients with FIP1L1-PDGFRA fusion are characterized by a male predominance, marrow fibrosis, increased number of mast cells, elevated serum tryptase levels, and a favorable response to low doses of imatinib. Most of these patients have a normal karyotype because a cryptic deletion of CHIC2 locus on 4q12 which is only 800 kb in size. This abnormality is detectable using a more sensitive FISH technology and RQ-PCR in the majority of cases. Moreover, the commercially available tricolor FISH probe will detect not only deletion 4q12 as a result of FIP1L1-PDGFRA but also a rare BCR-PDGFRA fusion resulting from the t(4;22)(q12;q11.2) rearrangement ( Fig. 57.23 ). Serial monitoring with RQ-PCR demonstrates exquisite sensitivity of FIP1L1-PDGFRA -positive patients to low-dose imatinib treatment ( Chapter 74 ). The FIP1L1-PDGFRA has also been detected in histopathologically defined cases of systemic mastocytosis with associated eosinophilia, and in cases of AML and T-cell lymphoblastic lymphoma associated with eosinophilia. In an Italian prospective cohort of 27 patients with FIP1L1-PDGFRA who were treated with imatinib, a complete hematologic response was achieved within 1 month and all patients became PCR-negative for FIP1L1-PDGFRA after a median of 3 months (range 1 to 10 months). In contrast to CML, very few cases of acquired imatinib resistance have been reported. Those rare reported cases had a mutation in T674I within the ATP-binding domain of PDGFRA, analogous to the T315I mutation in CML.
Although translocations involving PDGFRB at 5q32 region have been identified with 22 different fusion partners, they are indeed very rare. Any patient with a 5q31–q33 chromosomal abnormality with or without eosinophilia should be investigated for PDGFRB rearrangements by FISH testing ( Fig. 57.24 ).

Once the PDGFRA, PDGFRB , and FGFR1 rearrangements are excluded, the only other recurrent secondary abnormalities include trisomy 8 and trisomy 21 (see box on Genetic Testing for Ph-Negative Myeloproliferative Neoplasms ).
At diagnosis, perform a real-time alleles-specific polymerase chain reaction for JAK2V617F, as well as mutational studies for CALR and MPL. Cytogenetic analysis of marrow cells is recommended for patients with essential thrombocytopenia and polycythemia vera. Unstimulated peripheral blood can be used instead of marrow aspirate for patients with primary myelofibrosis patients. Perform FISH studies with BCR-ABL1 for patients with essential thrombocythemia to exclude the diagnosis of chronic myeloid leukemia. FISH studies can be performed when cytogenetics is uninformative for detection of the most frequent abnormalities: +1q, +8, +9, +9p, del(13)(q14), del(20)(q11q13). A panel of five probes includes CEP9, CDKN2A/B at 9p21, D8Z2 as a centromeric probe for chromosome 8, RB1 for deletion 13q, and D20S108 for deletion of 20q12. For patients with a normal karyotype perform aCGH+SNP testing because 68% of these patients have copy number abnormalities including cnLOH. For diagnostic purposes and to monitor treatment response in patients with hypereosinophilic syndrome, perform FISH using triple-color FIP1L1 probe. For 5q32 rearrangements perform FISH with a PDGFRB FISH probe.
In the WHO 2016 classification, mastocytosis is no longer listed as a Ph-negative MPN ( Chapter 56, Chapter 75 ) but rather as a distinct category, “Mastocytosis.” Mastocytosis includes a group of rare and heterogeneous diseases characterized by accumulation of clonal mast cells in one or multiple organs. Over 80% of patients with systemic mastocytosis ( Chapter 75 ) are characterized by KIT mutations, specifically in the activation loop of KIT , KIT D816V. KIT is encoded by a 21-exon-containing gene located on the long arms of chromosome 4, 4q12. Other rare KIT mutations have been described in adult and pediatric patients. The knowledge of the type and structure of KIT mutation is important because the A-loop mutations D816V/H/Y/N disrupt the structure of the receptor, leading permanently to an active conformation and resistance to imatinib. In patients with advanced systemic mastocytosis additional mutations have been observed including TET 2 (up to 39%), SRSF2 (up to 36%), ASXL1 (up to 21%), and RUNX1 (23%). At diagnosis of mastocytosis patients testing for KIT D816V in peripheral blood leukocytes, using highly sensitive and specific PCR-based assays, is necessary to determine the KIT allelic burden rapidly, which is now used as a gold standard for assessing disease burden.
The MDSs (see Chapter 61, Chapter 65 ) are a clinically heterogeneous group of hematologic neoplasms with differing biology and clinical manifestations. They have in common a clonal origin, dysplastic cellular morphology, cytopenias, abnormalities of cellular maturation, and an increased propensity to develop acute leukemia (20% to 40%). They predominantly occur in elderly people. Cytogenetic studies can provide both diagnostic and profound prognostic information regarding OS and the risk of transformation to AML. A chromosomally abnormal clone can be detected in 50% to 60% of patients with de novo MDS and in approximately 90% of patients with therapy-related MDS ( Table 57.5 ). There appears to be a correlation between the frequency of chromosomal abnormalities and the severity of disease. Approximately 35% of patients with less aggressive MDS, such as refractory anemia and refractory anemia with ring sideroblasts, have clonal chromosomal rearrangements, whereas approximately 60% to 70% of patients with refractory anemia with excess blasts in transformation have such chromosomal abnormalities. A single or complex chromosomal abnormality may be present initially, and cytogenetic progression may occur during the course of the disease. Even at diagnosis, complex genomic lesions involving five or more different chromosomes are not unusual. Despite the heterogeneity of chromosomal defects (gain, loss, deletion, amplification, rare balanced translocations, transcriptional silencing via methylation, or point mutation), the unifying concept of genetic instability in MDS is hemizygosity of specific genes or chromosomal regions. The heterogeneity of chromosomal abnormalities is overwhelming. However, the most common chromosomal anomalies observed in MDS include gain of 1q, del(5q)/−5, del(7q)/−7, trisomy 8, del(11)(q23), del(12p), +13/del(13q), t(11q23), del(17p), del(20)(q11q13), +21, and idic(X)(q13).
| Abnormality | Frequency (%) |
|---|---|
| −5 or del 5q | 10–15 |
| −7 or del 7q | 10 |
| trisomy 8 | 10–17 |
| i(17q) or t(17p) | 2–3 |
| del(12p) or t(12p) | 1–2 |
| del(11q) | 1–2 |
| −13 or del(13q) | 1–2 |
| del(9q) | 1–2 |
| idic(X) | 1 |
| inv(3)(q21q26.2) | 1 |
| t(6;9)(p23;q34) | 1 |
| t(3;21)(q26.2;q22.1) | <1 |
| t(1;3)(p36.3;q21.2) | <1 |
| t(11;16)(q23;p13.3) | <1 |
| t(2;11) (p21;q23) | <1 |
Until recently guidelines-based indicators (GBI) for MDS patient management did not receive adequate attention. Several population-based studies reported that approximately 25% of MDS patients do receive bone marrow cytogenetic analyses at diagnosis and the lack of chromosome analysis impeded appropriate risk stratification by IPSS-R. GBI indicators represent a standardized best practice performance, outcome, and clinically relevant guidelines for a patient with MDS. According to the diagnostic guidelines the initial work-up must include cytogenetic analysis and molecular diagnostics/NGS/ TP53 evaluation (among other parameters).
In 2012 a new cytogenetic classification of MDS was established that includes five different risk groups (see Chapter 61 ).
The clinical relevance and the power of conventional cytogenetics in MDS were recognized by the WHO, which documented a strong association between del(5)(q13q33) and 5q− syndrome. The 5q− syndrome is a unique subtype of low-risk MDS with a favorable prognosis, lack of other cytogenetic abnormalities, low risk of leukemic transformation, and more commonly occurs in older females ( Fig. 57.25 , second row ). Patients with isolated del(5q) remains the only MDS subtype defined by the genetic abnormality.
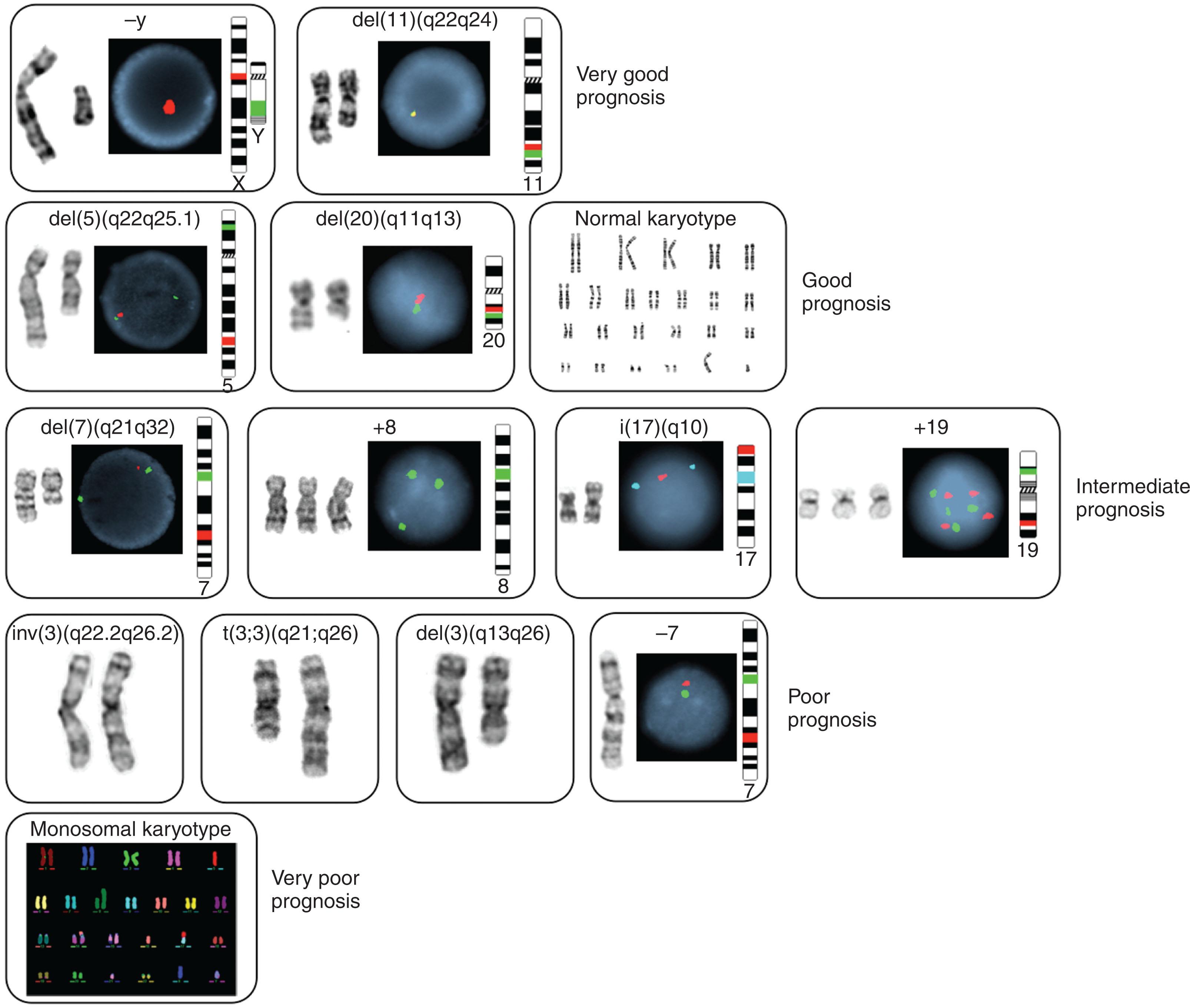
Among patients who do not have 5q− syndrome (with or without other chromosomal abnormalities), interstitial deletion of the long arms of chromosome 5 occur in 10% to 15% of patients and are among the most frequent chromosomal abnormalities in MDS (see Fig. 57.25 , second row ). The finding of del(5q) in CD34CD38− stem cells indicates its origin in a stem cell capable of differentiating into myeloid and lymphoid cell lineages. Data on 1432 patients with del(5q) showed a significant amount of heterogeneity in breakpoints. FISH studies delineated a commonly deleted segment that is currently estimated to be 1.5 Mb in size, on 5q31.1. The clustering of genes responsible for growth and differentiation of hematopoietic cells at this site and recurrent nature of −5/del(5q) in MDS caused many investigators to speculate that (a) tumor suppressor gene(s) was/were located in the 5q31 or 5q22–23 band region. To date, the tumor suppressor gene responsible for MDS on 5q has yet to be identified. Because the mechanism causing the interstitial del(5q) is elusive, haploinsufficiency or inactivation caused by methylation, rather than a typical tumor suppressor gene, has been speculated to be involved in this process.
Patients with isolated del(5q) have a more favorable prognosis and have a more prolonged survival than patients with ACAs. Specifically, patients with del(5)(q13q31) live longer than patients with other 5q deletions, indicating that the type of 5q deletion may significantly affect prognosis and response to therapy. Indeed, lenalidomide therapy leads to a normal karyotype in 44% of 148 patients with interstitial del(5q) ( Chapter 61 ). Isolated monosomy 7 or 7q deletion occurs in 20% of patients. Frequently, chromosome 7 abnormalities occur with ACA, most commonly rearrangements of 3q or del(12p) (see Fig. 57.25 , third and fourth row ). Monosomy 7 is present in all MDS subtypes and is seen predominantly in males. In pediatric patients with constitutional disorders associated with a predisposition to develop AML including Fanconi anemia (FA), congenital neutropenia, neurofibromatosis type 1, Down syndrome (DS), or Kostmann syndrome, −7/del(7q) may be seen as an isolated abnormality ( Chapter 30 ). Therefore the question remains to whether these patients have genetic imprinting and preferentially lose chromosome 7. Unequivocal evidence exists that preferential parental origin of the missing chromosome 7 does not occur; approximately half of the patients have loss of either the maternal or paternal homologue, excluding the genomic imprinting hypothesis. Embryonic origin of partial chromosome 7 deletion in monozygotic twins with juvenile chronic myelomonocytic leukemia has been reported. As shown in Fig. 57.26 , acquired cnLOH of 7q is a known recurrent genomic rearrangement in MDS patients not detected cytogenetically but identified using aCGH+SNP, and often detected in patients showing either a normal karyotype or other abnormalities.
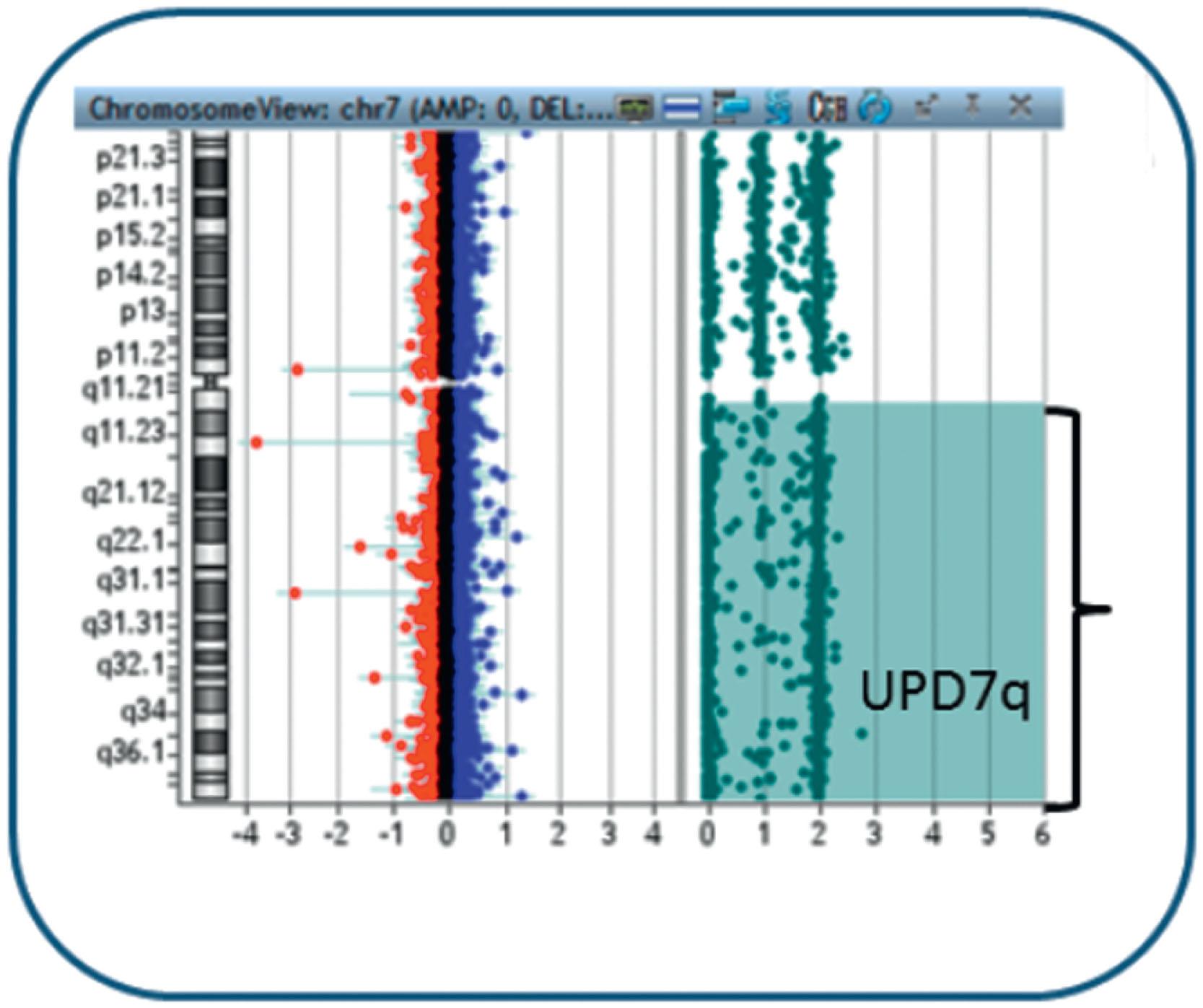
MDS patients with del(7q) as a single abnormality have a distinct clinicopathologic profile with an overall better prognosis than seen in patients with an isolated monosomy 7. Isolated del(7q) is more frequent in patients with less advanced forms of MDS according to the WHO classification or the IPSS-R. They have fewer blasts in the bone marrow than other cytogenetic groups and display a significantly superior survival when compared with patients with isolated monosomy 7. The presence of ACA in patients with del(7q) is associated with shortened OS (see Fig. 57.25 , third row ). Allele typing studies implicated three regions that are most frequently deleted: 7q22, 7q31.1, and 7q31.3. Isolated 7q deletion is a common finding in a variety of myeloid malignancies (MDS, AML, MPN, and others) and as a sole abnormality is present in less than 5% of patients. High-resolution genomic analysis has revealed that among 81 patients with a sole 7q deletion 67 cases had an interstitial deletion while 14 cases showed a terminal 7q deletion. In this cohort, according to the SNP array karyotyping, 85% of patients with interstitial 7q deletion had a commonly deleted regions located at 7q22.1 to 7q31.31. Consequently, there is speculation that a putative myeloid suppressor gene(s) is located in the regions that are frequently deleted. Because prototypic tumor suppressor genes have not been identified in patients with 7q deletions, an alternative explanation may be haploinsufficiency whereby the level of protein is critical, or a complex of two cooperating proteins is affected as a result of inactivation caused by methylation.
Trisomy 8 (see Fig. 57.25 , third row ) is the third most frequent chromosomal abnormality in MDS patients. As a sole abnormality, it is found in 11% of patients with MDS and overall is found in 17% of patients. A significantly higher incidence of trisomy 8 occurs in males than in females. Trisomy 8 is present in all age groups of patients with MDS. Although trisomy 8 is detected in the hematopoietic stem cells of patients with MDS, a sizable fraction of stem cells are disomic but functionally abnormal, suggesting that the acquisition of trisomy 8 is a secondary event. These findings provide evidence for the multistep pathogenesis of MDS, where gain of chromosome 8 is not an early event in the stepwise disease evolution. Although trisomy 8 serves as an intermediate risk factor when detected at diagnosis, patients with MDS with trisomy 8 treated with the hypomethylating agent 5-azacitidine have a significantly better survival than patients with other chromosomal abnormalities. Tetrasomy, pentasomy, and hexasomy 8 (polysomy 8) are relatively rare compared to trisomy 8. They have been reported in MDS, AML, or MPN. Tetrasomy 8 was the most common presentation of polysomy 8. In 60.7% of patients, polysomy 8 occurred as part of complex changes (16.2% with 11q23 rearrangements). No cryptic KMT2A rearrangements were found in cases when polysomy 8 was the only karyotypic change. Therefore polysomy 8 is a subtype of AML, MDS, and MPN characterized by a high incidence of prior diseases, and poor OS (6 months). Age significantly reduced median survival but associated cytogenetic abnormalities did not modify it. Cytogenetic results further demonstrate an in vitro preferential growth of the cells with a high level of aneuploidy suggesting a selective advantage for polysomy 8 cells.
MDS or MDS/MPN associated with trisomy 11 as a sole abnormality or as a part of noncomplex karyotype has been reported. This rare recurrent abnormality has an overall frequency of approximately 0.3% of MDS patients and is associated with a significantly inferior survival in patients with IPSS intermediate-risk disease ( P = .0002) but comparable to the poor-risk group ( P = .97). Trisomy 11 is associated with clinical aggressiveness and represents a high-risk cytogenetic abnormality.
Deletions of 17p are seen in 3% to 4% of MDS and AML patients. These patients often display several other chromosomal rearrangements, including monosomy 17, isochromosome 17q (see Fig. 57.25 , third row ), and unbalanced translocations between chromosome 17 and another chromosome. Approximately 30% of these deletions are related to therapy. The extent of 17p deletion in all cases involves the TP53 gene. There appears to be a close correlation between dysgranulopoiesis (e.g., pseudo–Pelger-Huet abnormalities) and small vacuoles in neutrophils with 17p abnormalities and TP53 deletion. The median survival of these patients is poor.
Besides deletion of TP53 , molecular characterization of MDS with NGS has shown that TP53 mutant MDS and therapy-related AML have emerged as molecular cohorts with profoundly inferior outcomes regardless of treatment, with a median OS of only 6 to 12 months. In MDS, mutations of TP53 are commonly seen in patients with complex karyotypes and have a significant negative impact on prognosis. TP53 mutations are strongly associated with thrombocytopenia, increased blasts, and chromosome 5 abnormalities, isolated del(5q), and complex karyotypes. Presence of TP53 mutations in patients with a complex karyotype identifies a patient group with an extremely poor prognosis and median OS of 6 months highlighting the unmet need for novel therapeutic approaches in these patients. Recent analysis of 3324 patients with MDS delineated two subsets of patients with distinct phenotypes and outcomes. One-third of TP53 -mutated patients had monoallelic mutations whereas two-thirds had multiple hits (multi-hit) consistent with biallelic targeting. Established associations with complex karyotype, few co-occurring mutations, high-risk presentation, and poor outcomes were specific for multi-hit patients only. The TP53 multi-hit state predicted risk of death and leukemic transformation independently of the IPSS-R. Surprisingly, monoallelic patients did not differ from TP53 wild-type patients in outcomes and response to therapy. These observations show that consideration of TP53 allelic state is critical for prognostication of MDS patients.
An isodicentric X chromosome in Xq13-idic(X) is a rare but recurrent abnormality that has been associated with refractory anemia with ringed sideroblasts. All MDS patients with idic(X)(q13) are females, most likely because the formation of idic(X) would result in nullisomy for Xq13-qter in males. The median age at the time of diagnosis is 73.5 years. The outcome of idic(X)-positive cases is variable; some investigators report aggressive and rapidly fatal disease and others a relatively favorable clinical course with survival for several years. The fact that idic(X) most often occurs as the sole cytogenetic abnormality suggests that it may be sufficient for leukemogenesis. Using a high-resolution SNP array, the breakpoints on idic(X)(q13) were mapped to two distinct breakpoint clusters, at approximately 70.9 Mb (five cases) and 72.1 Mb (seven cases) on the X chromosome. None of the 11 breakpoints occurred in a gene, strongly indicating that the idic(X)(q13) does not result in a fusion gene. Instead, the functional outcome of the abnormality confers a gene dosage effect because of the concurrent gain of Xpter-q13 and loss of Xq13-qter. This region of the X chromosome is enriched for repeated sequences and most likely, these repeats may facilitate the formation of idic(X). The isodicentric X chromosome was inactive in some patients and active in other patients; hence idic(X) appears to be leukemogenic regardless of X a or X i involvement.
Gain of 1q, usually in the form of an unbalanced translocation, is a recurrent abnormality in MDS and appears to be a marker of disease progression. Specifically, the gain of 1q in the form of jumping translocations either at diagnosis or the subsequent acquisition of jumping 1q translocations appears to be associated with imminent transformation to AML in patients after an average of 9 months ( Figs. 57.27 and 57.21 ). As mentioned earlier in the Ph-negative MPN section ( Chapter 70, Chapter 72 ), gain of 1q is also associated with disease progression and is most frequently found at the diagnosis of PV. Once acquired, the patient’s prognosis tended to be dependent on the copy number of 1q, indicating that gain of jumping 1q translocations was associated with both disease progression and poor prognosis. Jumping translocations of 1q has also been described in donor cell derived MDS after cord blood transplantation; the average time to developing jumping 1q was less than 2 years. These findings did not provide any common pattern as to the role of the partner chromosomes in unbalanced jumping 1q translocations. However, 81% of recipient breakpoints were localized in pericentric regions whereas the remaining 19% were telomeric fusions. Decondensation of pericentromeric heterochromatin of chromosome 1 together with centromere and repeat DNA sequences interspersed with histones and acetylated sequences may favor illegitimate recombinations leading to jumping 1q translocations. Moreover, exposure to azacitidine has been shown to be associated with alterations of pericentromeric heterochromatin of chromosome 1, as well as an increase in Alu DNA repeats. Hypomethylation of 1q12–21 pericentromeric region of chromosome 1 appears to be at least one aspect of copy number gains of 1q. The mechanism underlying formation of trisomy 1q in MDS has not been investigated but in patients with myelofibrosis we reported that +1q was associated with increased transcripts of MDM4 , likely leading to further downregulation of TP53 activity. Elevated MDM4 levels have been previously found in other hematologic malignancies, including de novo AML. There are numerous other oncogenes that are present on 1q, which could also contribute to disease progression. The overexpression of CKS1B , involved in cell cycle regulation, located at band 1q21 has been proposed as the culprit responsible for disease progression in patients with myeloma and gain of 1q ( Chapter 91 ). Therefore, it is possible that in MDS, similar to patients with myelofibrosis with +1q, leads to further dysregulation of the wild-type TP53 pathway by increasing MDM4 activity.
The most frequent rearrangement of chromosome 3 involves two bands on chromosome 3—band 3q21 and 3q26 simultaneously—which produces either t(3;3)(q21;q26) or inv(3)(q21q26) ( Fig. 57.28A and B ). These chromosomal rearrangements are present in de novo and therapy-related MDS, as well as in AML and megakaryoblastic crisis of CML. The incidence of the 3q rearrangements is 2% to 5%. Characteristic clinical features include an elevated platelet count, marked hyperplasia with dysplasia of megakaryocytes, and a poor prognosis with minimal or no response to chemotherapy and a short survival. In addition to similar clinicopathologic features, patients with 3q21q26 share molecular heterogeneity in both the breakpoints and the expression pattern of the genes near these breakpoints (see Fig. 57.28 , also Fig. 57.30A .) The chromosomal breakpoints, defined by FISH, in 3q26 are scattered over several hundred kB in either the 5′ or the 3′ region of the MECOM ( EVI1 complex locus) gene, whereas the breakpoints in the 3q21 region are restricted to two different smaller genomic clusters approximately 100 kb downstream of the RPN1 gene. EVI1 overexpression is observed in the majority of patients, but some patients with the 3q21q26 rearrangement do not have detectable EVI1 expression, and at least 9% of patients with AML without 3q26 abnormalities overexpress EVI1 . Therefore the poor prognosis of these patients may be independent of EVI1 expression, despite the fact that extensive 3q26 breakpoint FISH mapping suggests EVI1 involvement in numerous novel sporadic and recurrent 3q26 rearrangements. A fusion transcript of RPN1-EVI1 is rarely observed in patients with 3q21q26 rearrangements. Interestingly, functional genomic studies and allelic-specific analysis revealed experimentally that inv(3)/t(3q) results simultaneously in haploinsufficiency of GATA2 and upregulation of MEECOM as a result of rearrangements in noncoding regulatory sequences of these genes, indicating the importance of mutations residing outside the coding exome.
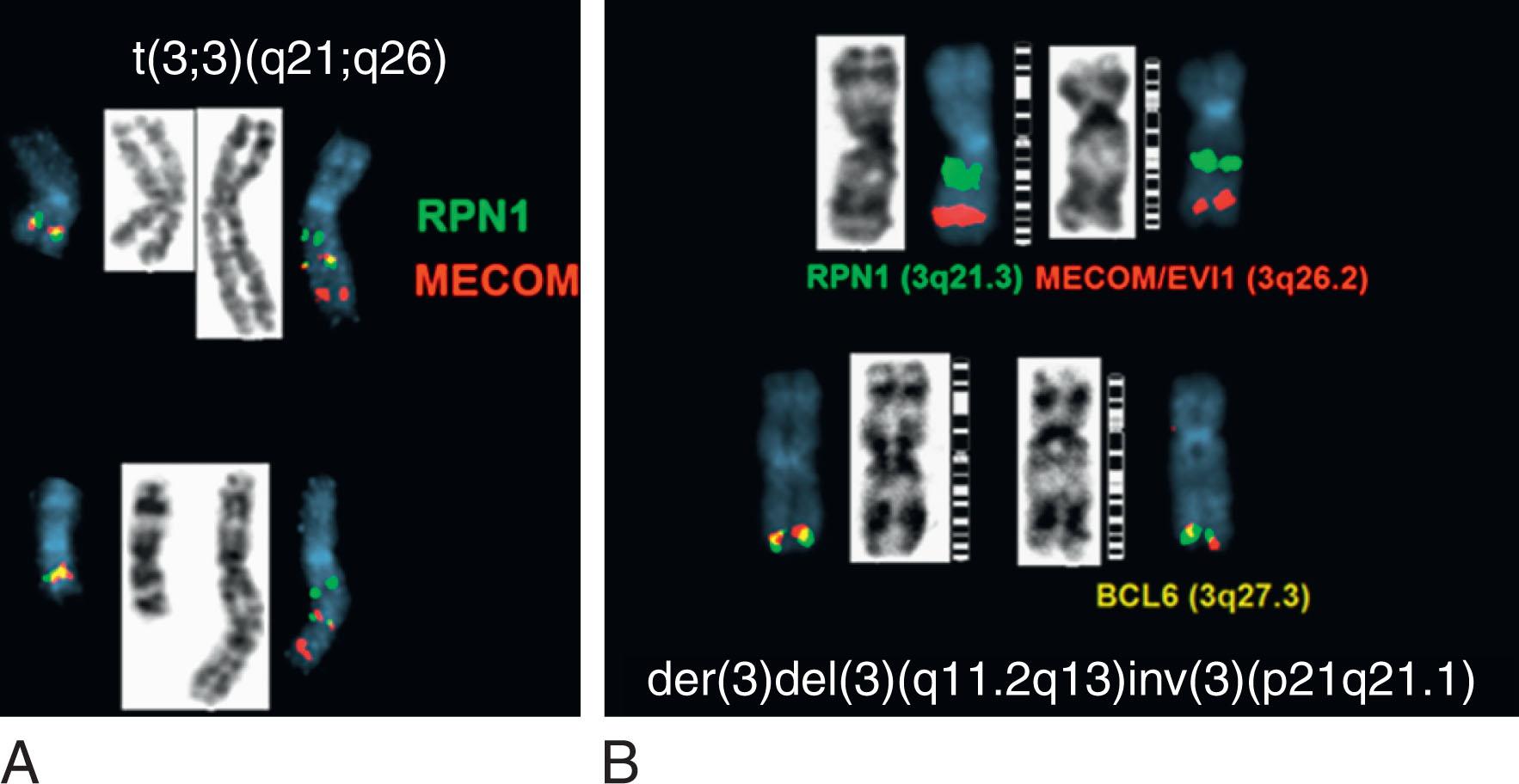
A monosomal karyotype (MK), a new cytogenetic category, is defined as a karyotype showing two or more distinct autosomal chromosome monosomies or one single autosomal monosomy (excluding isolated loss of X or Y) in the presence of a structural abnormality (see Fig. 57.25 , bottom row ). Initial reports indicated that MDS with a MK, with or without monosomy for chromosomes 5 and 7, was associated with a poorer prognosis than other complex karyotypes, although later reports could not confirm these results. The reason for this discrepancy may be due to azacitidine treatment with reducing the negative impact of MK in high-risk patients with MDS.
The clinical significance of LOY present in 10% of patients with MDS and approximately 7% of older adult males without MDS was recently elucidated (see section above on LOY in clonal hematopoiesis). Mosaic LOY in blood has been reported to be associated with higher risk of all-cause mortality and overall lower life expectancy. In bone marrow samples containing more than 75% of metaphase cells with a LOY, the observation is associated with MDS. Structural Y chromosome abnormalities are also present in patients with MDS ( Fig. 57.29 ). Older adult males with MDS and LOY who achieve complete hematologic remission regain the Y chromosome in their marrow cells.
The prognosis of patients with MDS is very heterogeneous. In 1997, based on the cytogenetic abnormalities identified in 816 patients with MDS, as well as percentage of blasts and number of cytopenia, an IPSS was proposed. According to the IPSS, 86% of all cytogenetic findings can be explicitly classified according to their prognostic impact. The system is highly reproducible and very simple to use, but it has certain limitations. Moreover, in the remaining 14% of patients the cytogenetic abnormalities have unknown prognostic significance ( Fig. 57.30 ). This limitation underscores two major limitations of cytogenetic classifications of MDS: the profound heterogeneity of acquired cytogenetic aberrations in MDS and the associated challenge of designing a comprehensive cytogenetic scoring system that predicts the prognostic impact of rare abnormalities ( Fig. 57.31 ). A new and comprehensive cytogenetic scoring system based on an international data collection of 2902 patients was reported in 2012 (see Fig. 57.25 ). Patients included were from the German-Austrian MDS Study Group (n = 1193) , the International MDS Risk Analysis Workshop (n = 816) , the Spanish Hematological Cytogenetics Working Group (n = 849) , and the International Working Group on MDS Cytogenetics (n = 44) databases. In total, cytogenetic categories were defined, providing clear prognostic classification for 91% of all patients. All abnormalities were arranged according to OS and development of AML to classify their prognostic impact. The abnormalities were classified into five prognostic subgroups: very good ( n = 81) included del(11q) and LOY (median OS, 61 months); good (n = 1809) included normal karyotype, del(5q), del(12p), and del(20q) (all as single anomaly) and double abnormalities including del(5q) (median OS 49 months); intermediate (n = 529) included del(7q), +8, i(17q)(q10), +19, +21, any other single abnormality, independent clones, and double abnormalities not harboring del(5q) or −7/del(7q) (median OS 26 months); poor (n = 148) included inv(3)/t(3q)/del(3q), −7, and double abnormalities including −7/del(7q) and complex (three abnormalities; OS of 16 months); and very poor (n = 197) included complex karyotypes with more than three abnormalities (OS of 6 months) (see Fig. 57.25 . This new scoring system proposed should be viewed as a dynamic model, open to further refinement as the knowledge in karyotypic abnormalities of MDS evolves.
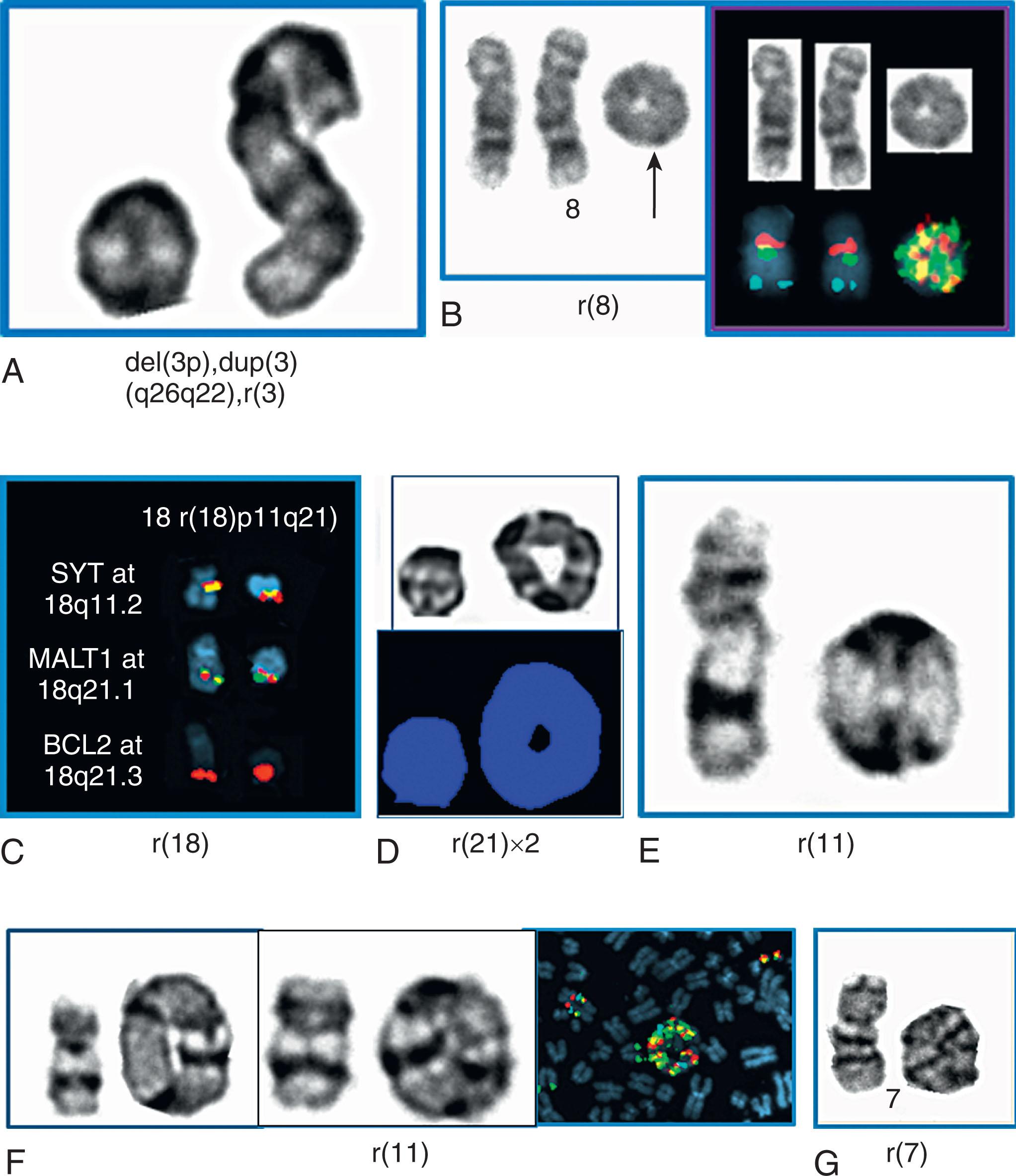
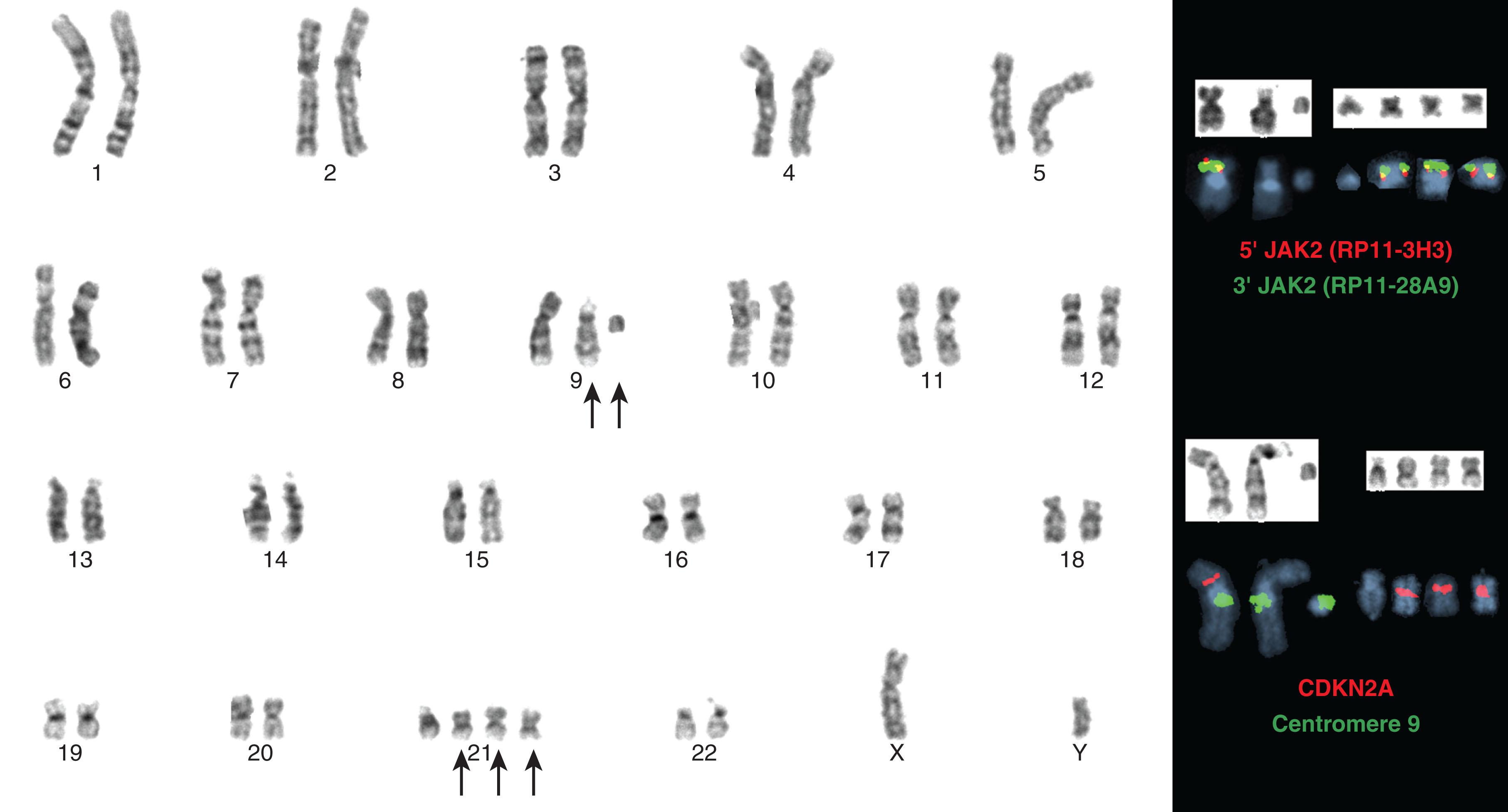
Cytogenetics and FISH studies have relatively similar sensitivities in detecting an abnormal clone among patients with MDS. A FISH test for targeted recurrent abnormalities in MDS should use probes to detect numerical and structural anomalies of chromosome regions 1q, 3q, 5q, 7, 7q31, 8, 11q, 12p, 13q, 17p, 20q, and 21 (see box on Genetic Testing for Myelodysplastic Syndrome and Fig. 57.25 ).
Due to the tremendous chromosomal and genomic heterogeneity the best genetic test at diagnosis is conventional cytogenetic studies. It is also required for the IPSS-R prognosis. FISH for detection of P53 single or biallelic deletion will identify a group of patients with very poor prognosis. In patients with a normal karyotype a reflex test is aCGH+SNP for detection of cnLOH, specifically on 7q, allowing greater confidence in detection of genomic change fostering improved patient-specific management.
Detection rate of submicroscopic or cryptic aberrations in MDS patients with normal karyotype is 32.8%. The chromosomal abnormalities detected by SNP-A can influence the prognosis of patients with MDSs, especially in the low-risk group. The patients with multiple chromosomal abnormalities detected by SNP karyotyping had an inferior prognosis, and those with SNP abnormalities (≥3 per patient) were found to be an independent predictor of poor prognosis in patients with MDS. Moreover, SNP karyotyping in MDS revealed chromothripsis (chromosomal shattering) as recurrent genomic abnormality (1.2%), specifically in high-risk MDS cases, and the presence of chromosome 13 chromothripsis is a novel recurrent genomic change in high-risk MDS.
Familial MDS is very rare ( Chapter 30, Chapter 61 ). MDS presenting in children and younger adults is more frequently associated with germline genetic predisposition ( Chapter 65 ). In recognition of the growing subgroup of MDS or acute leukemia associated with germline mutations or familial cases, the revised WHO of myeloid neoplasms now includes a new category: myeloid neoplasms with germline predisposition ( Chapter 30 ). The diagnosis of a germline MDS predisposition disorder carries profound implications for medical management and treatment. Although there is a whole spectrum of germline MDS predisposition conditions a few of those associated with acquired chromosomal abnormalities will be listed here. MDS with germline GATA2 mutations is frequently associated with monosomy 7/del7q (−7) or trisomy 8, particularly in children and young adults. A study of 426 cases of pediatric MDS patients identified germline GATA2 mutations in 37% of patients with primary MDS with −7 and 16% of MDS cases with trisomy 8 ( Chapter 65 ). Germline GATA2 mutations were identified in 72% of adolescents with −7 MDS. In contrast, germline GATA2 mutations were absent in therapy-related MDS. Germline mutations in SAMD9 and SAMD9L genes are frequently present in patients with bone marrow failure and a high risk for monosomy 7 MDS at a young age and these patients present either with MDS or AML. FA is characterized by genomic instability, hypersensitivity to DNA cross-linking agents, and predisposition to marrow failure, as well as hematologic malignancies and solid tumors ( Chapter 30 ). The cumulative incidence of patients with FA to develop MDS is 40% by age 50 years and 20% by age 40 years for AML ( Chapter 30 ). The incidence of leukemia is even higher in the FANCD1/BRCA2 subtype of FA, with many cases presenting at ages less than 5 years. In the registry of patients with Diamond-Blackfan anemia (DBA) of North America, 2% of DBA patients developed AML by age 45 years. Dyskeratosis congenita (DC) or telomere biology disorders (TBDs) encompass genetically heterogeneous disorders related to impaired telomere maintenance ( Chapter 30 ). These patients often present associated with hematologic complications including bone marrow failure, MDS, and AML. The cumulative incidence of MDS in patients with DC has been estimated to be 2% by age 50 years. RUNX1 deficiency is the cause of an autosomal dominant familial platelet disorder with predisposition to myeloid leukemia. Familial platelet disorder with predisposition to developing MDS, AML, or lymphoid malignancies such as T-cell leukemia or B-cell malignancies ( Chapter 30 ). Another constitutional abnormality that appears to predispose to MDS/AML is a deletion of band region q22.1 to q22.2 of chromosome 21 (where RUNX1 gene is located) with a complex phenotype (dysmorphic features, organ malformations, growth delay, and mental retardation), germline RUNX1 deletion, and congenital thrombocytopenia. Although longitudinal data are limited, lifetime risk for developing a myeloid malignancy in patients with germline RUNX1 mutations appears to be highly variable and has been estimated to be around 44%, ranging from 11% to 100%. Tables 57.6 and 57.7 shows the acquired recurrent somatic mutations observed in patients with MDS. According to the European Leukemia Net report 52% to 90% of patients with a normal karyotype have at least one genomic point mutation or MDS-related copy number change as detected by combing three technologies: NGS, cytogenetics, and a CGH+SNP. Using SNP array data from 50,000 subjects recruited for genome-wide association studies demonstrated that 0.5% of healthy individuals possessed genomic lesions. In contrast, recent whole-exome sequencing data of DNA from the peripheral blood of 17,182 persons showed clonal mutations leading to clonal outgrowth of hematopoietic cells in 10% of persons over the age of 70 years and in 18.4% among people aged 90 to 108 years. The most commonly mutated such genes were DMT3A , TET2 , and ASXL1 , which are frequent mutations in MDS and AML. These elderly people with clonal mutations in their peripheral blood did not have MDS or any other hematologic malignancy, indicating that age-related CH is a common premalignant condition and individuals with clonal mutations have an increased risk of developing hematologic malignancy ( Chapter 13, Chapter 19 ).
| Gene | Chromosomal Location | Frequency (%) | Prognosis |
|---|---|---|---|
| SFRB1 | 2q33.1 | 25–30 | Favorable? |
| TET2 | 4q24 | 20–25 | Neutral |
| RUNX1 | 21q22.12 | 10–20 | Unfavorable |
| ASXL1 | 20q11.21 | 10–15 | Unfavorable |
| SRSF2 | 17q25.1 | 10–15 | Unfavorable |
| TP53 | 17p13 | 5–10 | Unfavorable |
| U2AF1 | 19q13.42 | 5–10 | Unfavorable |
| NRAS/KRAS | 1p13.2/12p12.1 | 5–10 | Unfavorable |
| DNMT3A | 2p23.3 | 5 | Unfavorable |
| ZRSR2 | Xp22.2 | 5 | Neutral? |
| EZH2 | 7q35-36 | 5 | Unfavorable |
| IDH1/IDH2 | 2q33.3/15q26.1 | 2–3 | Unfavorable |
| ETV6 | 12p13 | 2 | Unfavorable |
| CBL | 11q23.3 | 1–2 | Unfavorable |
| NPM1 | 5q35.1 | 1–2 | ? |
| JAK2 | 9p24 | 1–2 | Unfavorable |
| SETBP1 | 18q12.3 | 1–2 | ? |
| SF3A1 | 22q12.2 | 1–2 | ? |
| SF1 | 11q13.1 | 1–2 | ? |
| U2AF65 | 19q13.42 | 1–2 | Unfavorable |
| PRPF40B | 2q23.3 | 1–2 | ? |
| RNA Splicing | |
| SF3B1 | 20%–35% |
| SRSF2 | 10%–15% |
| U2AF1 | 5%–10% |
| ZRSR2 | <5% |
| DNA Methylation | |
| TET2 | 11%–26% |
| DNMT3A | 7%–10% |
| Histone Modification | |
| ASXL1 | ∼20% |
| EZH2 | ∼5% |
| DNA Transcription | |
| RUNX1 | ∼10% |
| TP53 a | ∼5% |
| Signal Transduction | |
| NRAS | <5% |
| KRAS | <5% |
| PTPN11 | <5% |
| Chromatid Cohesion | |
| STAG2 | <10% |
| RAD21 | <5% |
| SMC1A | <5% |
| SMC3 | <5% |
The widespread use of NGS has identified more than 50 recurrent somatic mutations in patients with MDS leading to the development of a rich mosaic detailing the mutational landscape and genetic determinants of prognostic significance and survival in MDS ( Chapter 3, Chapter 13, Chapter 19, Chapter 61 ). The only mutation that has diagnostic validity is an acquired mutation in SF3B1 gene which was recognized by WHO for its potential to establish diagnosis of MDS with ring sideroblasts. Four lines of evidence support recognition of somatic SFRB1 mutation as a disease-defining genomic event: (1) its most often an initial (founding) genetic event occurring in the most primitive hematopoietic stem cells (CD34+CD38−CD90+CD45RA-cells), (2) it is a major determinant of the disease phenotype, (3) it has an independent prognostic parameter which influences survival and risk of progression to AML, and (4) it may predict response to specific agents. With the widespread adoption of targeted sequencing panels it is now recognized that more than 80% of MDS patients harbor one or more known recurrent mutation. TP53 VAF (variable allele frequency) may inform distinct risk subgroups because of lower-risk patients with TP53 mutations at the time of diagnosis, more than 6% VAF were linked to shorter OS and progression-free survival compared to patients with less than 6% VAF.
AML refers to a group of heterogeneous diseases with respect to clonality, molecular lesions, chromosomal aberrations, and response to treatment (see Chapter 59, Chapter 60, Chapter 63 ). Initially, using G6PD as a marker of clonality, it was determined that AML originates from a single clone and has a multistep pathogenesis. In adults, at the time of diagnosis all hematopoietic cell lineages are clonal. In children younger than 16 years, erythroid cells and platelets often are not part of the leukemic clone.
The application of nucleotide-polymorphism technologies has confirmed the clonal origin of AML and further revealed that most, if not all, patients with AML evolve through a process of constrained clonal evolution. Multicolor metaphase analysis, FISH, CGH arrays, SNP arrays, PCR, and NGS have all greatly improved detection of subclones and established the concept of a clonal hierarchy in AML. To understand the branching evolution of AML development, each mutation must be placed in the context of whether it is a “driver” versus “passenger” and an “initiation” versus “progression” event ( Chapter 59 ). Driver mutations directly affect the biology of the cell, whereas passenger mutations do not. Initiation mutations are present within the founding clone and are found in all AML cells. Progression mutations emerge during leukemic evolution, leading to subclones, and exist in only a fraction of AML cells. Selection pressure, such as chemotherapy, can favor the elimination or outgrowth of different branches within the AML evolutionary hierarchy.
Modern genomic technologies have provided a better understanding of normal and leukemic hematopoiesis ( Chapter 3 ). A subset of AML cases evolve from a preceding clinically overt phase such as MDS by subclonal evolution with an increased number of genomic changes. The founder mutations present in preleukemic cells are retained in AML blasts, implicating them as putative initiating events and establishing clonal expansion as the first step in leukemogenesis. However, some recurrent leukemia-associated somatic mutations, such as TET2 , have also been linked to multilineage CH in aging healthy individuals. For the majority of de novo AML without any prior clinical symptoms, the cell of origin, biologic consequences of initiating mutations, and order of subsequent mutations remain poorly understood. Today, state-of the-art AML diagnostics relies on integration of different disciplines including cytogenetics and molecular genetics to provide most optimal diagnosis and management. Initial observations by the International Workshop on Chromosomes in Leukemia asserted that cytogenetic findings may serve as the single most important prognostic marker in AML; these observations were subsequently validated through studies conducted by the Medical Research Council (MRC), the Cancer and Leukemia Group B (CALGB), and the Southwest Oncology Group. Finally, in 2008, the WHO incorporated cytogenetic findings along with morphology, immunologic markers, and molecular genetics into their classification system ( Chapter 56 ). Cytogenetics remains the most powerful independent prognostic factor and provides the framework for risk stratification schemes that have been generally adopted to guide treatment approaches for AML patients. Based on karyotype status, two major groups of AML can be distinguished: (a) those with an abnormal karyotype, which accounts for approximately 52% of patients, and (b) those with a normal karyotype by conventional cytogenetics, which accounts for 48% of patients with AML.
AML is generally considered a medical emergency implying an immediate start of treatment as soon as the diagnosis is made. However, a report from the German Study Alliance Leukemia-Acute Myeloid Leukemia registry investigated 2263 patients and analyzed the relationship between the time from diagnosis to treatment start (TDT) and concluded that the TDT had no prognostic significance on remission rates and OS. They suggested that it may be safe to wait for genetic and other laboratory test results in order to assign patients to the best available treatment option before the start of intensive therapy for AML.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here