Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Our understanding of the origins of cancer has changed dramatically over the past three decades, due in large part to the revolution in molecular biology that has altered the face of all biomedical research. Powerful experimental tools have been thrust into the hands of cancer biologists. These tools, including newly devised and implemented technologies that permit the interrogation of entire genomes, have made it possible to uncover and dissect the complex molecular machinery operating inside the single cell, normal and malignant, to understand its operations, and to pinpoint the defects that cause cancer cells to proliferate abnormally.
Three decades ago, at least three rival models of cancer’s origins had substantial following among those interested in the roots of cancer. One model portrayed cancer as a disease of abnormal differentiation. According to this thinking, the changes in cell behavior that occur during the process of development run awry during tumor progression, causing cells to make inappropriate choices in moving up or down differentiation pathways. This concept of cancer’s origins had important implications for the molecular origins of cancer: because the process of differentiation involves changes in cell phenotype without underlying changes in the genome, this model suggested that cancer was essentially an epigenetic process—a change in cell behavior without an underlying change in its genetic constitution.
An alternative model was advanced by the virologists. By the early 1970s, a number of distinct cancer-causing viruses had been catalogued in various animal species and in humans. These ranged from the Rous sarcoma virus, whose discovery reached back to the first decade of the century, to Shope papillomavirus, Epstein-Barr virus, papovaviruses such as SV40 and polyomavirus, and a variety of retroviruses that infected various mammals and birds. The existence of these viruses suggested that similar agents operated to trigger human tumors. Such hypothetical human tumor viruses were thought capable of insinuating themselves into human cells and transforming them from a normal to a malignant growth state.
Yet another way of explaining cancer’s origins was advanced by those who were impressed by the increasing connections being forged between carcinogens and mutagens. More than half a century of experiments had demonstrated the abilities of radiation as well as a vast array of chemicals to induce tumors in animals and occasionally in humans. Independent of this research, Drosophila and bacterial geneticists had documented the abilities of some of these carcinogenic agents to act as mutagens. The most influential of these experiments was to come from the laboratory of Bruce Ames. In the mid-1970s, Ames described a correlation between the mutagenic potencies of various chemical compounds and their respective potencies to induce tumors in laboratory animals.
Ames’ correlation ( Figure 1-1 ) yielded the inference that the carcinogenic powers of agents derive directly from their abilities to damage genes and thus the DNA of cells. This strengthened the convictions of those who had long embraced the notion that cancer cells were really mutants and that their abnormal behavior derived from mutant genes that they carried in their genomes. This model implied that such mutant genes arose through somatic mutations, i.e., mutations that occur in somatic tissues during the lifetime of an organism and alter genes that were pristine at the moment of conception.
This last model of cancer’s origins would eventually dominate thinking; the other two models largely fell by the scientific wayside. As the 1970s progressed, the search for tumorigenic viruses associated with most types of common human cancers bogged down. Human papillomavirus (HPV) clearly had strong associations with cervical carcinomas, Epstein-Barr virus (EBV) with Burkitt’s lymphomas in Africa and nasopharyngeal carcinomas in southeast Asia, and hepatitis B and C viruses (HBV, HCV) with hepatocellular carcinomas in east Asia. Together, these accounted for as much as 20% of tumors worldwide. However, the remaining types of cancers, and thus the vast majority of human cancers arising in the Western world, had no obvious viral associations in spite of extensive attempts to uncover them.
The epigenetic model of cancer lost its attractiveness largely because an extensive array of mutant growth-controlling genes was discovered in the genomes of human tumor cells. So the focus shifted increasingly to genes, more specifically the genomes of cancer cells. Cancer genetics in the 1970s and early 1980s became a branch of somatic cell genetics—the genetics of cells and their somatically mutated genes. Indeed, advances in the technology of DNA sequencing have now enabled the enumeration of mutations present in specific cancer genomes and will eventually lead to a compendium of recurrent genetic alterations in human cancers.
The notion that cancer cells were mutants should have motivated a systematic search for genes that suffered mutation during the development of tumors. Moreover, these mutant genes should possess another property: they needed to specify some of the aberrant phenotypes ascribed to tumor cells, including alterations in cell shape, decreased dependence on external mitogenic stimuli, and an ability to grow without tethering to a solid substrate (anchorage independence). The fact that viruses were not important causative agents of most types of human tumors generated another conclusion about these cancer-causing genes: they were likely to be endogenous to the cell rather than being imported into the cell from some external source. Stated differently, it seemed likely that these cancer genes were mutant versions of preexisting normal cellular genes.
In the 1970s, when this line of thinking matured, the experimental opportunities to test its validity were limited. The human genome, which harbored these hypothetical cancer genes, represented daunting complexity. Its vastness precluded any simple, systematic survey strategy designed to locate mutant growth-controlling genes within cancer cells. Indeed, it is only now, three decades later, that the means, deep sequencing of cancer genomes, for conducting effective systematic surveys for cancer genes has been developed. Thus the discovery of cancer-causing genes—oncogenes as they came to be called—depended on a circuitous, indirect experimental strategy.
Ironically, it was tumor viruses, in the midst of being discredited as important etiologic agents of human cancer, that led the way to finding the elusive cancer genes. Varmus and Bishop’s study of the Rous sarcoma virus (RSV) broke open the puzzle. Their initial agenda was to understand the replication strategy of this chicken virus. However, in the years after 1974, they focused their attentions to unraveling the mechanism used by RSV to transform an infected normal cell into a tumor cell.
Earlier work of others had indicated that a single gene, named src, carried the vital cancer-causing information present in the viral genome. Accordingly, the Varmus and Bishop laboratory launched a research program to trace the origins of this virus-associated src oncogene. In fact, the origins of most viral genes were obscure, shrouded in the deep evolutionary past. It seemed that most viruses and thus their genes originated hundreds of millions of years ago, perhaps as derivatives of the cells that they learned to parasitize.
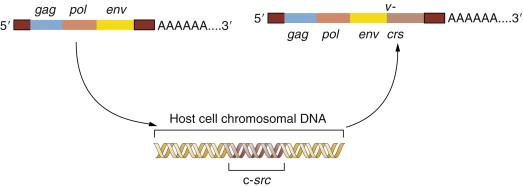
However, as this team reported in 1976, the src gene behaved differently: it was a recent acquisition by the Rous virus. Many closely related retroviruses shared with RSV an ability to replicate in chicken cells and a very similar set of genes needed for viral replication. However, these other viruses lacked the src gene and the ability to transform infected cells into cancer cells, suggesting that the src oncogene carried by RSV was a relatively recent genetic acquisition. The Varmus-Bishop group soon traced the origins of the src gene to an unexpected source—a closely related gene that resided in the genome of normal chickens and, by extension, in the genomes of all vertebrates. They named this gene c- src (cellular src ) to distinguish it from the v- src (viral src ) oncogene carried by the virus.
The Varmus-Bishop evidence converged on a simple conceptual model. It explained all their observations and ultimately much more. The progenitor of RSV lacked the v- src gene but grew well in chicken cells. During one of its periodic forays into a chicken cell, this ancestor virus picked up a copy of the c- src gene and incorporated it into its own viral genome. Once src was present within the viral genome, this slightly remodeled gene—now v- src —was exploited by RSV to transform cells it encountered in subsequent rounds of infection.
This provided a testimonial to the cleverness and plasticity of retroviruses, which seemed able to capture and then exploit normal cellular genes to do their bidding. But another implication was even more important: the Varmus-Bishop work pointed to the existence of a normal cellular gene, the c- src gene, that seemed to possess a latent ability to induce cancer. This cancer-causing ability was unmasked when the c- src gene was abducted by the chicken retrovirus that became the progenitor of RSV ( Figure 1-2 ).
The c -src gene was named a proto-oncogene to indicate its inherent potential to become activated into a cancer-causing oncogene. Within several years, it became clear that as many as a dozen other tumorigenic retroviruses also carried oncogenes, each of which had been abstracted from the genome of an infected vertebrate cell. Hence, there were many proto-oncogenes in the normal cell genome, not just c- src. Each seemed to be present in the DNA of a normal mammalian or avian host species, and by extension, present as well in the genomes of all vertebrates.
These discoveries were momentous because they demonstrated that normal cellular genes had the ability to induce cancer if removed from their normal chromosomal context and placed under the control of one or another retrovirus. Still, a key piece was missing from this puzzle. Retroviruses seemed to be absent from most, indeed from almost all, human tumors. Could proto-oncogenes ever become activated without direct intervention by a marauding retrovirus?
An obvious response was that proto-oncogenes might be altered by mutational events that did not remove these genes from their normal chromosomal roosts. Instead, these mutations would alter proto-oncogenes in situ in the chromosome by affecting either the control sequences or the protein-encoding sequences of these genes. This notion led to another question: If some proto-oncogenes could become activated by somatic mutations, such as those inflicted by chemical or physical carcinogens, would these be the same proto-oncogenes that were the targets of mobilization and activation by retroviruses?
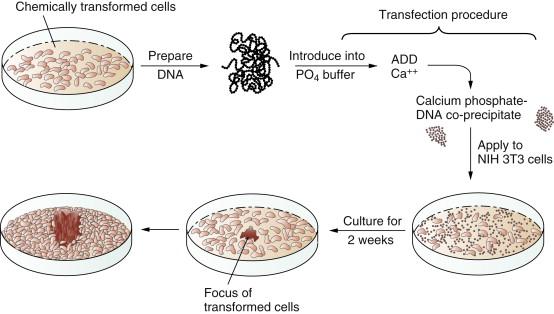
In 1979 and 1980, answers came, once again from unexpected quarters. These newer experiments depended on the use of gene transfer, also known as transfection. The transfection procedure could be used to convey DNA, and thus genes, from tumor cells into normal recipient cells. The goal here was to see whether the transferred tumor cell DNA could induce some type of malignant transformation in the recipient cells. Success in such an experiment would indicate that the transferred gene(s) previously operated in the donor tumor cell to induce its transformation.
These transfection experiments succeeded ( Figure 1-3 ). DNA extracted from chemically transformed mouse fibroblasts was able to induce normal mouse fibroblasts to undergo transformation. Retroviruses were clearly absent from both the donor tumor cells and the recipients that underwent transformation and so could not be invoked to explain the cancer-causing powers of the transferred DNA. Soon the identity of these transferred genes, which functioned as oncogenes, became apparent. They were members of the ras family of oncogenes, which had initially been discovered through their association with rodent sarcoma viruses. These rodent retroviruses had acquired ras proto-oncogenes from normal rodent cells, much like RSV, which had stolen a copy of the src proto-oncogene from a chicken cell.
Unanswered by this was the genetic mechanism that imparted oncogenic powers to the tumor-associated ras oncogene, more specifically an H- ras oncogene. It soon became clear that the tumor-associated H- ras oncogene was closely related to, indeed virtually indistinguishable from, a normal H- ras proto-oncogene that was present in the genomes of all vertebrates. Still, the tumor-associated ras oncogene carried different information than did the precursor proto-oncogene: the oncogene caused the malignant transformation of cells into which it was introduced, whereas the counterpart proto-oncogene had no obvious effects on cell phenotype. This particular puzzle was solved in 1982 with the finding that an H- ras oncogene cloned from a human bladder carcinoma carried a point mutation—a single nucleotide substitution—that distinguished it from its counterpart proto-oncogene. This genetic alteration, clearly a somatic mutation, sufficed to convert a normally benign proto-oncogene into a virulent oncogene.
Within months, yet other activated oncogenes were found in human tumors by using DNA probes prepared from a variety of retrovirus-associated oncogenes. The myc oncogene, initially associated with avian myelocytomatosis virus, was found to be present in increased gene copy number (i.e., amplified) in some human hematopoietic tumors ; in yet others, myc was activated through a chromosomal translocation that juxtaposed its coding sequences with those of immunoglobulin genes, thereby placing the expression of the myc gene under the control of these antibody genes rather than its own normal transcriptional control elements. These discoveries extended and solidified a simple point: a common repertoire of proto-oncogenes could be activated either by retroviruses (usually in animal tumors) or by somatic mutations (in human tumors). The activating mutations involved either base substitution, amplification in gene copy number, or chromosomal translocation.
The discoveries of mutant, tumor-associated oncogenes in human tumors led to a simple model of cancer formation. Mutagenic carcinogens entered into cells of a target tissue and mutated a proto-oncogene. The resulting oncogene then induced the now-mutant cell to initiate a program of malignant growth. Eventually, years later, the progeny of this mutant founder cell formed a large enough mass to become a macroscopically apparent tumor.
While satisfying conceptually, this simple model of cancer formation clearly conflicted with a century’s worth of histopathologic analyses, which had indicated that tumor formation is really a multistep process, in which initially normal cell populations pass through a succession of intermediate stages on their way to becoming frankly malignant. Each of these intermediate stages contains cells that were more aberrant than those seen in the preceding steps. This body of observations persuaded many that the formation of a malignancy depended on a succession of phenotypic changes in the cells forming these various growths. Quite possibly, each of these shifts in cell phenotype reflected a change in the underlying genetic makeup of the evolving pre-malignant cell population. Such a multistep genetic model of tumor progression stood in direct conflict with the single-hit model of transformation that was suggested by the discovery of the point-mutated ras oncogene.
By 1983, one solution to this dilemma became apparent. In that year, experiments showed that a single introduced oncogene could not transform fully normal rat cells into ones that were tumorigenic. Two and maybe even more oncogenes seemed to be required to effect this conversion. For example, whereas an introduced ras oncogene could not transform normal embryo cells into tumor cells, the co-introduction of a ras plus a myc oncogene, or a ras plus an adenovirus E1A oncogene, succeeded in doing so. It appeared that such pairs of oncogenes collaborated with one another to induce the full malignant transformation of normal cells ( Figure 1-4 , A ). Moreover, this experiment suggested that human tumors carried two or more mutant oncogenes that collaborated with one another to orchestrate the many aberrant phenotypes associated with highly malignant cells.
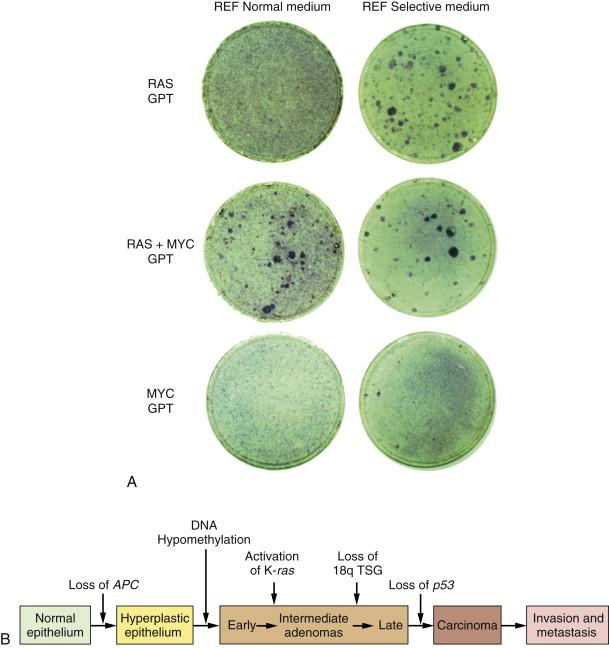
Observations such as these pointed to a new way of conceptualizing the multistep tumorigenesis long studied by the pathologists. It seemed plausible that each of the histopathological transitions arising during tumor development occurred as a consequence of a new mutation sustained in the genome of an evolving, premalignant cell population ( Figure 1-4 , B ). According to this thinking, tumor development was a form of Darwinian evolution, in which each successive mutation in a growth-controlling gene conferred increased proliferative potential and thus selective advantage on the cells bearing the mutant gene. Ultimately, a multiply mutated cell bearing half a dozen or more mutant genes might exhibit all of the phenotypes associated with highly malignant cancer cells.
This mechanistic model was validated through the creation of transgenic mice. Cloned copies of mutant oncogenes, such as ras and myc, were introduced into the germlines of mice. These transgenes were structured so that the oncogene was placed under the control of a transcriptional promoter that ensured expression of the resulting “transgene” in a specific tissue or developmental stage. Now the presence of a mutant oncogene in a particular tissue could be guaranteed through the actions of an appropriately engineered transgene rather than being dependent on the random actions of mutagenic carcinogens.
In one highly instructive group of experiments, a myc or a ras oncogene was placed under the control of the mouse mammary tumor virus transcriptional promoter, which guaranteed its expression in the mammary epithelium of the pregnant female mouse. As anticipated, these mice contracted breast cancer at extremely high rates. This demonstrated that mutant oncogenes were far more than markers of cancer progression; indeed, they could actually play a causal role in driving tumor pathogenesis.
Significantly, the transgenic mice did not contract cancer rapidly in their mammary tissue even though a mutant oncogene was implanted and expressed in virtually all of the epithelial cells of their mammary glands. Instead, their mammary carcinomas arose with several months’ delay, indicating that a second (and perhaps third) alteration was required in addition to the activated transgene before mammary epithelial cells launched a program of malignant growth. The nature of this additional alteration(s) was not always clear, but it almost certainly involved stochastic somatic mutations striking the mammary epithelial cells, creating mutant growth-controlling genes that collaborated with the transgene to trigger the outgrowth of malignant cell clones. In the years that followed, this work was extended to many types of human tumors, the cells of which were found to possess multiple mutant genes that contributed to tumor formation.
The model of multistep tumorigenesis implied that a tumor cell carries two or more mutant oncogenes, each activated by somatic mutation during one of the stages of tumor development. However, experimental validation of this model initially proved to be difficult. Most attempts at detecting mutant oncogenes in human tumor genomes yielded a ras or perhaps a myc oncogene, but rarely were two mutant oncogenes found to coexist in the genomes of human tumor cells. This left two logical alternatives. Either the genome of a typical human tumor cell did not contain multiple mutated genes, as the multistep model of cancer suggested, or there were indeed multiple mutated cancer-causing genes in tumors, but many of these were not oncogenes of the type that had been studied intensively in the 1970s and early 1980s.
In fact, there were candidate genes waiting in the wings. These others operated in a fashion diametrically opposite to that of the oncogenes: they seemed to prevent cancer rather than favoring it and came to be called “tumor suppressor genes.” Several independent lines of evidence led to the discovery and characterization of these genes.
Experiments using cell hybridization initiated by Henry Harris in Oxford provided the first indication of the existence of these suppressor genes. These cell hybridizations involved the physical fusion of two distinct types of cells that were propagated in mixed cultures. The conjoined cells would form a common hybrid cytoplasm and ultimately pool their chromosomes, yielding a hybrid genome.
Often these cell hybridizations involved the fusion of cells with two distinct genotypes. In some of these experiments, tumor cells were fused with normal cells. The motive here was to see which genome would dominate in determining the behavior of the resulting hybrids. Counter to the expectations of many, the resulting hybrid cells turned out, more often than not, to be nontumorigenic. This indicated that the genes present in the normal genome dominated over those carried in the cancer cell. In the language of genetics, the normal alleles were dominant, whereas the cancer cell–associated alleles were recessive. (More properly, the alleles present in the cancer cell created a phenotype that was recessive to the normal cell phenotype.)
This unanticipated behavior could most easily be rationalized by assuming that normal cells carried certain growth-normalizing genes, the presence of which was needed to maintain normal proliferation. Cancer cells seemed to have lost these genes, ostensibly through mutations that resulted in inactivated versions of the genes present in normal cells. When reintroduced into the cancer cells via cell fusion, the normal alleles reimposed control on the cancer cells, restoring their behavior to that of a normal cell. In effect, these growth-normalizing genes suppressed the tumorigenic phenotype of the cancer cells and were, for this reason, termed tumor suppressor genes (TSGs).
In their normal incarnations, the TSGs seemed to constrain growth, unlike the proto-oncogenes, which seemed to be involved in promoting normal proliferation. Inactivated, null alleles of TSGs were found in tumor cell genomes in contrast to the hyperactivated alleles of proto-oncogenes (i.e., oncogenes) found in these genomes.
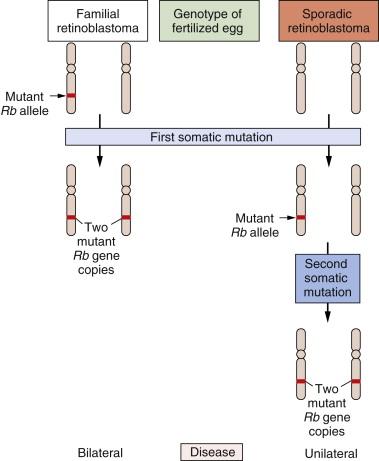
The study of retinoblastoma, the childhood eye tumor, converged on these cell hybridization studies in a dramatic way. This work had been pioneered by Alfred Knudson, who, beginning in the early 1970s, studied the genetics of this rare tumor. Knudson learned much by comparing the two forms of this cancer: sporadic retinoblastoma, which seemed to be due exclusively to accidental somatic mutations, and familial retinoblastoma, which appeared, like many familial cancers, to be due to the transmission of a mutated gene in the germline.
Knudson’s analysis of the kinetics of retinoblastoma onset persuaded him that a common set of gene(s) operated to generate both kinds of tumors. Although the nature of these genes eluded him, their number was clear. Sporadic retinoblastomas seemed to arise following two successive somatic mutations affecting a lineage of cells in the retina. The triggering of familial retinoblastomas seemed to require only a single somatic mutation. Knudson speculated that in these familial tumors, a second mutated gene was required to trigger tumorigenesis and that this gene was already present in mutant form in all the cells of the retina, having been inherited in mutated form from a parent of the affected child.
For the cancer geneticist, Knudson’s most important concept was the notion that a retinal cell needed to lose two mutant genes before it was transformed into a tumor cell. Sometimes one of the two mutant null alleles was contributed by the germline; more often, both genes arose through somatic mutation. However, the nature of these genes and the mutations that recruited them into the tumorigenic process remained elusive. Finally, in 1979, karyotypic analysis of a retinoblastoma revealed an interstitial deletion in the q14 band of chromosome 13. Later work revealed that this resulted in the loss of a gene, termed RB . Hence, one of the two mutational events needed to make a retinoblastoma involved the inactivation of an RB gene copy, in this particular case through the wholesale deletion of the chromosomal region carrying the RB gene.
By 1983, the nature of the second mutational event became clear: it involved the loss of the second, hitherto intact copy of the RB gene. Hence, the two mutational events hypothesized by Knudson involved the successive inactivation of the two copies of this gene. Suddenly, the need for two mutations became clear: The first mutation left the cell with a single, still-intact copy of the RB gene, which was able, on its own, to continue programming normal proliferation. Only when this surviving gene copy was eliminated from the cell genome did runaway proliferation begin ( Figure 1-5 ). Thus, mutations that inactivate an RB gene copy create alleles that function recessively at the cellular level. Only when both wild-type alleles are lost through various mutational mechanisms does a retinal cell begin to behave abnormally.
The RB gene became the paradigm for a large cohort of similarly acting TSGs that suffer inactivation during tumor progression. These TSGs are scattered throughout the cell genome and act through a variety of cell-physiologic mechanisms to control cell proliferation. They are united only by the fact that they control proliferation in a negative way, so that their loss permits uncontrolled cell multiplication to proceed.
The discovery of the RB gene gave substance and specificity to the genes that Harris had postulated from his cell fusion experiments. Equally important, they opened the door to understanding a variety of familial cancer syndromes. In the case of RB , inheritance of a mutant, defective allele predisposes to retinoblastoma early in life with more than 90% probability. Inheritance of a defective allele of the APC TSG predisposes with high frequency to adenomatous polyposis coli syndrome and thus to colon cancer. The presence of a mutant TP53 gene in the germline leads to increased rates of tumors in a number of organ sites, including sarcomas and carcinomas, yielding the Li-Fraumeni syndrome. More than two dozen heritable cancer syndromes have been associated with germline inheritance of defective TSGs.
In each case, the inheritance of a mutant, functionally defective TSG allele obviates one of two usually required somatic mutations. Because an inactivating somatic mutation represents a low-probability event per cell generation, the presence of an already-mutant inherited TSG allele enormously accelerates the overall kinetics of tumor formation. As a consequence, the likelihood of a tumor arising during the course of a normal lifespan is enormously increased.
The search for TSGs has been difficult, as their existence only becomes apparent when they are absent from a cellular genome. However, one peculiarity of TSG genetics has greatly aided the discovery of these genes. This involves the genetic mechanisms by which the second copy of a TSG is lost. In principle, two independent somatic mutations could successively inactivate the two copies of a TSG, thereby liberating a cell from the growth-constraining influences of this gene. However, each of these mutations normally occurs with a low probability—perhaps 10 −6 per cell generation. The likelihood of both mutations occurring is therefore roughly 10 −12 per cell generation, an extremely low probability. (Actually, because cancer cell genomes become progressively destabilized as tumors develop, this probability is usually higher.)
In fact, evolving premalignant cell populations carrying a single, already-inactivated TSG copy often resort to another genetic mechanism to eliminate the second, still-intact copy of this TSG. They discard the chromosomal arm (or chromosomal region) carrying the still-intact TSG copy and replace it with a duplicated copy of the chromosomal region carrying the mutant, already-inactivated TSG copy. All this is achieved via the exchange of genetic material between paired homologous chromosomes.
The end result of these genetic gymnastics is the duplication of the mutant TSG copy. Thus, the TSG goes from a heterozygous state (involving one mutant and one wild-type gene allele) to a homozygous state (involving two mutant gene copies). Almost always, the chromosomal region flanking the TSG suffers the same fate. Consequently, known genes as well as other genetic markers within this flanking region that were initially present in a heterozygous configuration now become reduced to a homozygous configuration. This genetic behavior has motivated cancer geneticists to analyze the genomes of human tumor cells, looking for chromosomal regions that repeatedly suffer loss of heterozygosity (LOH) during tumor progression. Such LOHs represent presumptive evidence for the presence of TSGs in these regions whose second wild-type copies have been eliminated by LOH during the course of tumor development. Once such a region is localized to a chromosomal region, several currently available gene molecular strategies can be exploited to further narrow the chromosomal domain carrying the TSG and ultimately to isolate the TSG through molecular cloning.
The existence of many dozen still-unknown TSGs is suspected because of the documented LOH affecting specific chromosomal regions of various types of human tumor cells. The effort to identify and clone these genes is being greatly facilitated by efforts such as those included in the International Cancer Genome Consortium and the Cancer Genome Anatomy Project (TCGA). Nonetheless, the successful identification and cloning of a significant cohort of TSGs has already provided one solution to a major puzzle posed earlier. As mentioned, although human tumor cells were hypothesized to carry a number of distinct, mutated growth-controlling genes, most tumors appeared to carry only a single activated oncogene. We now realize that many of the other targets of mutation during tumor progression are TSGs. Their inactivation collaborates with the activated oncogenes to create malignant cells and thus tumors. In the widely cited study of human multistep tumor progression—that described in colonic tumors by Vogelstein and his co-workers—the mutation of a K- ras oncogene is accompanied by mutations of the APC and TP53 TSGs and a third TSG that maps to chromosome 18. This evidence, together with a wealth of genetic studies reported subsequently, indicates that TSGs are inactivated even more frequently than oncogenes are activated during the course of forming many types of human tumors. Importantly, the inactivation of TSGs often phenocopies the cell-biological effects of oncogenes. This means that the inactivation of TSGs is as important to the biology of tumor progression as oncogene activation.
Unexpectedly, the discovery of TSGs also made it possible to understand how a variety of DNA tumor viruses succeed in transforming the cells that they infect. Unlike retroviruses, these DNA viruses carry oncogenes that have resided in their genomes for millions, and likely hundreds of millions, of years. Any connections with antecedent cellular genes, to the extent they once existed, were obscured long ago by the extensive remodeling that these oncogenes underwent while being carried in the genomes of the various DNA tumor viruses. Independent of their ultimate origins, it was clear in the 1980s that the oncogenes (and encoded oncoproteins) were deployed by DNA viruses to perturb key components of the normal cellular growth-controlling circuitry. However, the precise control points targeted by these viral oncoproteins remained obscure.
In the late 1980s, it was learned that a number of DNA tumor virus oncoproteins bind to the products of two centrally important TSGs, pRB and p53. For example, the large T oncoprotein of SV40 binds and sequesters both the p53 and pRB proteins of infected host cells; the E6 and E7 oncoproteins of human papillomaviruses target p53 and pRB, respectively. As a consequence, a virus-infected cell is deprived of the services of these two key negative regulators of its proliferation. Indeed, these virus-mediated inactivations closely mimic the state seen in many nonviral tumors that have been deprived of pRB and p53 function by somatic mutations striking the TSGs specifying these two proteins. So the transforming mechanisms used by these viruses could be rationalized by referring to the same genes and proteins that were known to be inactivated by mutational mechanisms in many types of spontaneous, nonviral human tumors. Importantly, these findings reinforced the notion that a single, central growth-regulating machinery operating in all types of cells suffers disruption by a variety of ostensibly unrelated genetic mechanisms, leading eventually to the formation of cancers.
The activation of oncogenes and the loss of TSGs together explain many of the phenotypes that one associates with cancer cells. These cells are able to grow without attachment to solid substrate, the aforementioned phenotype of anchorage independence, and they are able to grow on top of one another, which is manifested in culture as the loss of contact inhibition. Moreover, when compared to normal cells, cancer cells exhibit a greatly reduced dependence on mitogens and an ability to resist the antiproliferative effects of growth-inhibitory signals, such as those conveyed by transforming growth factor-β (TGF-β). Alterations of oncogenes and tumor suppressor genes can be invoked to explain these neoplastic cell traits.
Arguably the most interesting trait of cancer cells is their ability to resist a variety of stimuli and stresses that would cause normal cells to activate the cell-suicide program termed apoptosis. The fact that virtually all tumor cells have developed various types of resistance to apoptosis indicates that severe pro-apoptotic stresses are experienced repeatedly as normal cells evolve progressively toward a malignant phenotype and that an ability to resist these stresses is strongly selected during this evolution. Thus, changes in the complex array of genes that control entrance in the apoptotic program are frequently demonstrable within tumor cells. Although these genes are specialized in regulating a discrete cancer cell phenotype (apoptosis), they behave operationally like oncogenes and TSGs, i.e., the activation of some of these confers a resistance to apoptosis as does the inactivation of yet others.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here