Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The shapes of mammalian cells vary widely depending on their interactions with each other, their extracellular environment and internal structures. Their surfaces are often highly folded when absorptive or transport functions take place across their boundaries. Cell size is limited by rates of diffusion, either that of material entering or leaving cells, or that of diffusion within them. Movement of macromolecules can be much accelerated and also directed by processes of active transport across the plasma membrane and by transport mechanisms within the cell. According to the location of absorptive or transport functions, apical microvilli ( Fig. 1.1 ) or basolateral infoldings create a large surface area for transport or diffusion.
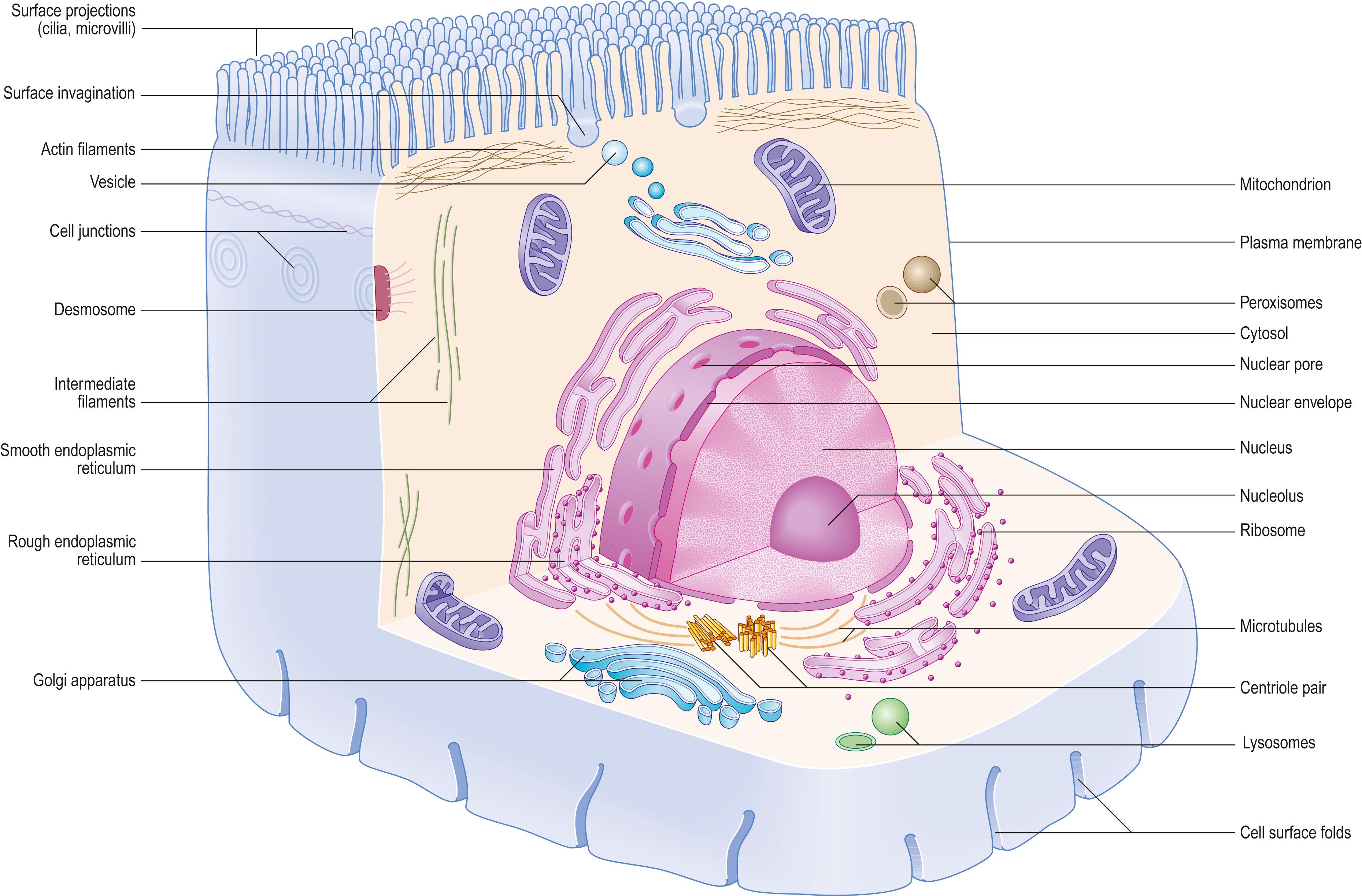
Motility is a characteristic of most cells, in the form of movements of cytoplasm or specific organelles from one part of the cell to another. It also includes: the extension of parts of the cell surface such as pseudopodia, lamellipodia, filopodia and microvilli; locomotion of entire cells, as in the amoeboid migration of tissue macrophages; the beating of flagella or cilia to move the cell (e.g. in spermatozoa) or fluids overlying it (e.g. in respiratory epithelium); cell division; and muscle contraction. Cell movements are also involved in the uptake of materials from their environment (endocytosis, phagocytosis) and the passage of large molecular complexes out of cells (exocytosis, secretion).
Epithelial cells rarely operate independently of each other and commonly form aggregates by adhesion, often assisted by specialized intercellular junctions. They may also communicate with each other, either by generating and detecting molecular signals that diffuse across intercellular spaces, or more rapidly by generating interactions between membrane-bound signalling molecules. Cohesive groups of cells constitute tissues, and more complex assemblies of tissues form functional systems or organs.
Most cells are between 5 and 50 μm in diameter: e.g. resting lymphocytes are 6 μm across, red blood cells 7.5 μm and columnar epithelial cells 20 μm tall and 10 μm wide (all measurements are approximate). Some cells are much larger than this: e.g. megakaryocytes of the bone marrow and osteoclasts of the remodelling bone are more than 200 μm in diameter. Neurones and skeletal muscle cells have relatively extended shapes, some of the former being over 1 m in length.
Each cell is contained within its limiting plasma membrane, which encloses the cytoplasm. All cells, except mature red blood cells, also contain a nucleus that is surrounded by a nuclear membrane or envelope (see Fig. 1.1 ; Fig. 1.2 ). The nucleus includes: the genome of the cell contained within the chromosomes; the nucleolus; and other subnuclear structures. The cytoplasm contains cytomembranes and several membrane-bound structures, called organelles, which form separate and distinct compartments within the cytoplasm. Cytomembranes include the rough and smooth endoplasmic reticulum and Golgi apparatus, as well as vesicles derived from them. Organelles include lysosomes, peroxisomes and mitochondria. The nucleus and mitochondria are enclosed by a double-membrane system; lysosomes and peroxisomes have a single bounding membrane. There are also non membrane-bound structures, called inclusions, which lie free in the cytosolic compartment. They include glycogen aggregates and pigments (e.g. lipofuscin). In addition, ribosomes and several filamentous protein networks, known collectively as the cytoskeleton, are found in the cytosol. The cytoskeleton determines general cell shape and supports specialized extensions of the cell surface (microvilli, cilia, flagella). It is involved in the assembly of specific structures (e.g. centrioles) and controls cargo transport in the cytoplasm. The cytosol contains many soluble proteins, ions and metabolites.
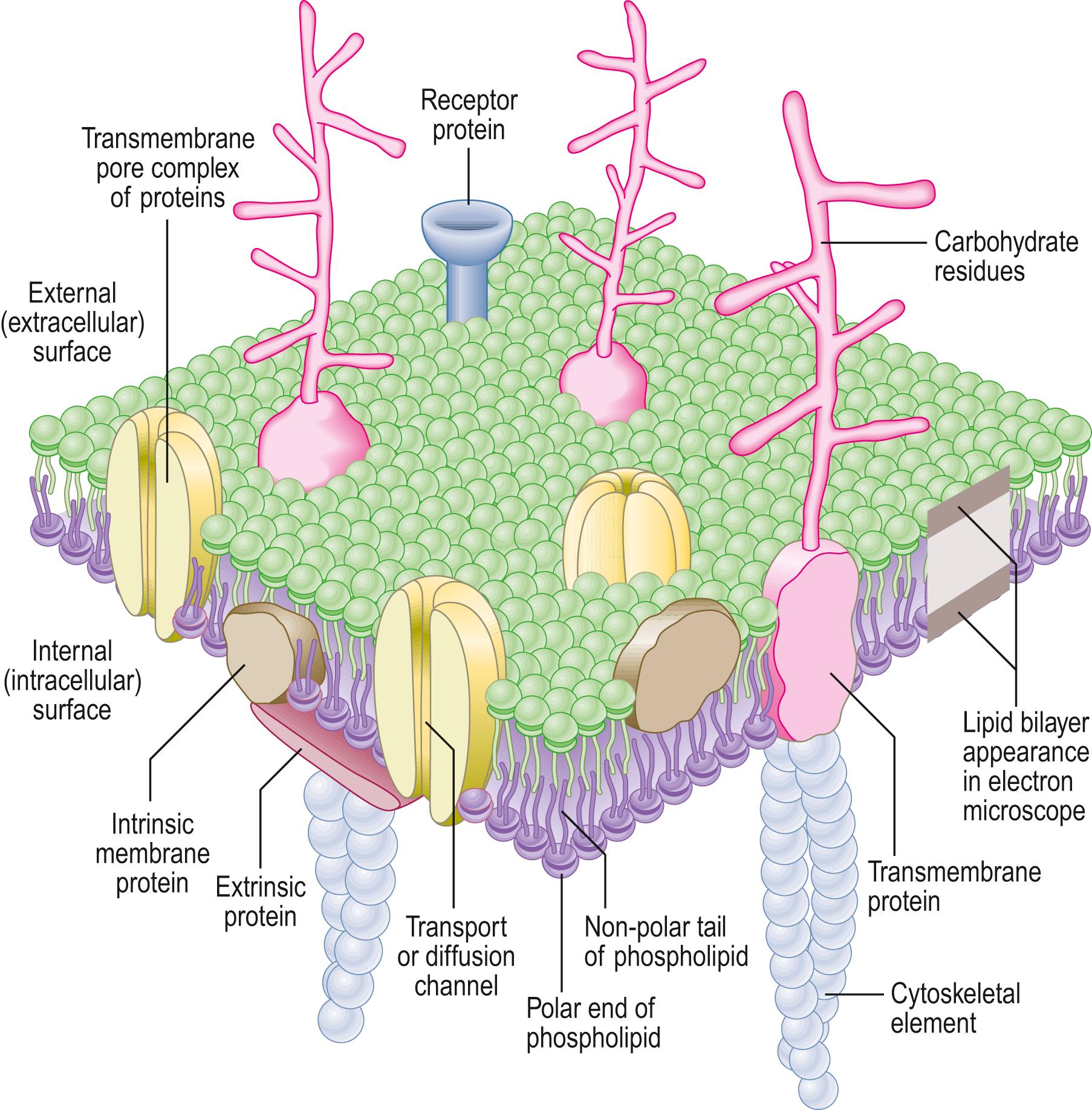
Cells are enclosed by a distinct plasma membrane, which shares features with the cytomembrane system that compartmentalizes the cytoplasm and surrounds the nucleus. All membranes are composed of lipids (mainly phospholipids, cholesterol and glycolipids) and proteins, in approximately equal ratios. Plasma membrane lipids form a lipid bilayer, a layer two molecules thick. The hydrophobic ends of each lipid molecule face the interior of the membrane and the hydrophilic ends face outwards. Most proteins are embedded within, or float in, the lipid bilayer as a fluid mosaic. Some proteins, because of extensive hydrophobic regions of their polypeptide chains, span the entire width of the membrane (transmembrane proteins), whereas others are only superficially attached to the bilayer by lipid groups. Both are integral (intrinsic) membrane proteins, as distinct from peripheral (extrinsic) membrane proteins, which are membrane-bound only through their association with other proteins. Carbohydrates in the form of oligosaccharides and polysaccharides are bound either to proteins (glycoproteins) or to lipids (glycolipids), and project mainly into the extracellular domain ( Fig. 1.3 ) .
Combinations of biochemical, biophysical and biological techniques have revealed that lipids are not homogeneously distributed in membranes, but that some are organized into microdomains in the bilayer, called ‘detergent-resistant membranes’ or lipid ‘rafts’, rich in sphingomyelin and cholesterol. The ability of select subsets of proteins to partition into different lipid microdomains has profound effects on their function, e.g. in T-cell receptor and cell–cell signalling. The highly organized environment of the domains provides a signalling, trafficking and membrane fusion environment.
In the electron microscope, membranes fixed and contrasted by heavy metals such as osmium tetroxide appear in section as two densely stained layers separated by an electron-translucent zone – the classic unit membrane. The total thickness of each layer is about 7.5 nm. The overall thickness of the plasma membrane is typically 15 nm. Freeze–fracture cleavage planes usually pass along the hydrophobic portion of the bilayer, where the hydrophobic tails of phospholipids meet, and split the bilayer into two leaflets. Each cleaved leaflet has a surface and a face. The surface of each leaflet faces either the extracellular surface (ES) or the intracellular or protoplasmic (cytoplasmic) surface (PS). The extracellular face (EF) and protoplasmic face (PF) of each leaflet are artificially produced during membrane splitting. This technique has also demonstrated intramembranous particles embedded in the lipid bilayer; in most cases, these represent large transmembrane protein molecules or complexes of proteins. Intramembranous particles are distributed asymmetrically between the two half-layers, usually adhering more to one half of the bilayer than to the other. In plasma membranes, the intracellular leaflet carries most particles, seen on its face (the PF). Where they have been identified, clusters of particles usually represent channels for the transmembrane passage of ions or molecules between adjacent cells (gap junctions).
Biophysical measurements show the lipid bilayer to be highly fluid, allowing diffusion in the plane of the membrane. Thus proteins are able to move freely in such planes unless anchored from within the cell. Membranes in general, and the plasma membrane in particular, form boundaries selectively limiting diffusion and creating physiologically distinct compartments.
Most biological molecules cannot diffuse through the phospholipid bilayer. Specific transport proteins, such as carrier proteins and channel proteins, mediate the selective passage of molecules across the membrane, thus allowing the cell to control its internal composition. Molecules (such as oxygen and carbon dioxide) can cross the plasma membrane down their concentration gradients by dissolving first in the phospholipid bilayer and then in the aqueous environment at the cytosolic or extracellular side of the membrane. This mechanism, known as passive diffusion, does not involve membrane proteins. Lipid and lipid-soluble (e.g. steroid) substances can also cross the bilayer.
Other biological molecules (such as glucose, charged molecules and small ions H+, Na+, K+ and Cl–) are unable to dissolve in the hydrophobic interior of the phospholipid bilayer. They require the help of specific transport proteins ( Fig. 1.4 ) and channel proteins, which facilitate the passage or facilitated diffusion of most biological molecules. Facilitated diffusion requires carrier proteins, which can bind specific molecules to be transported, or channel proteins, which form open gates through the membrane. Carrier proteins transport sugars, amino acids and nucleosides. Channel proteins are ion channels involved in the rapid transport of ions (faster transport than that mediated by carrier proteins), are highly selective of molecular size and electrical charge, and are not continuously open. Ligand-gated channels open ‘gates’ in response to the binding of a signalling molecule, whereas voltage-gated channels open in response to changes in electric potential across the membrane. Similar to passive diffusion, facilitated diffusion of biological molecules is determined by concentration and electrical gradients across a membrane.
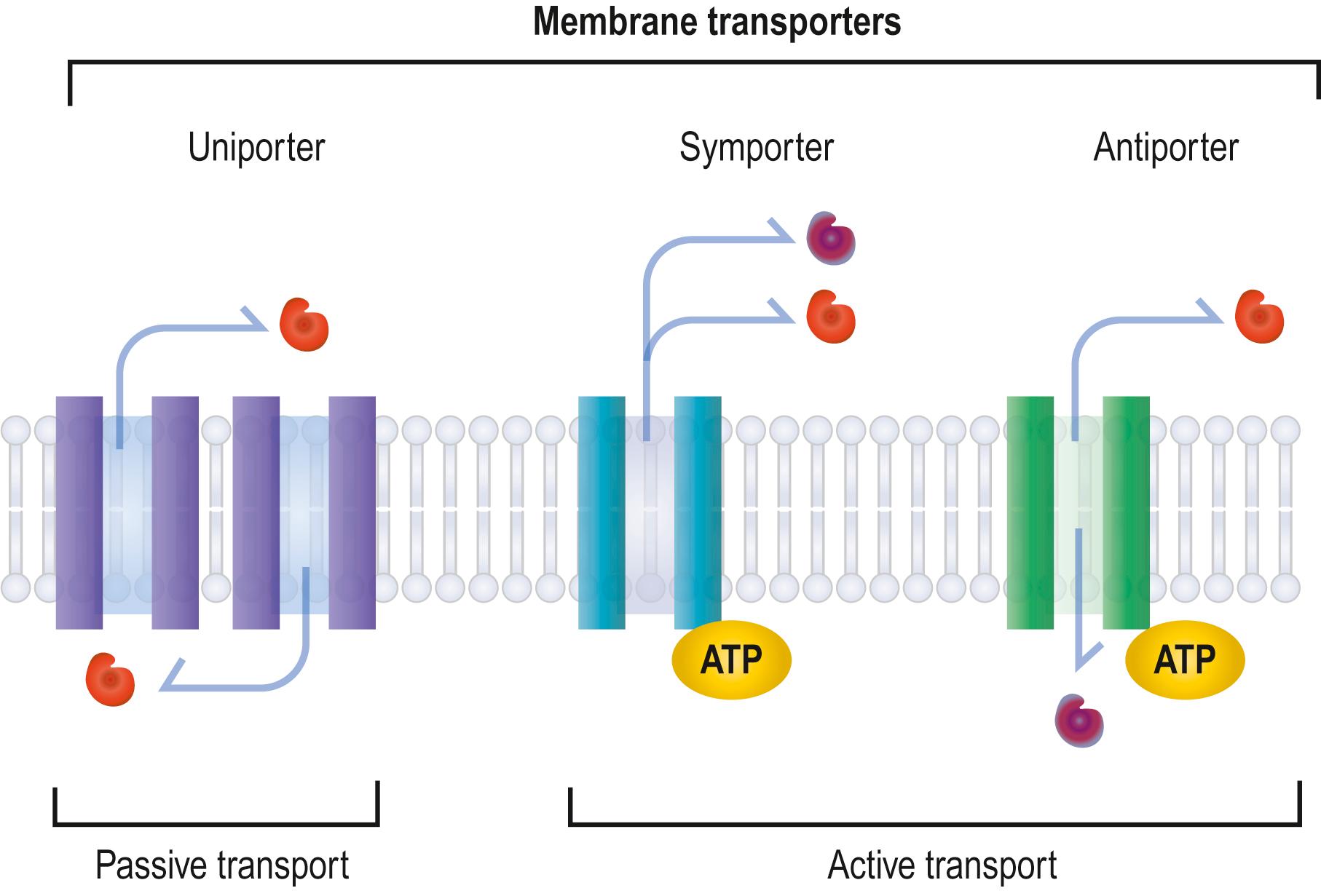
There are three types of transport protein: uniporter, symporter, and antiporter (see Fig. 1.4 ). Uniporter transport proteins can be either channel proteins or carrier proteins. They are involved in facilitated diffusion and carry a single molecule at a time from one side of the membrane to the other along its concentration gradient. Symporter and antiporter transport proteins are involved in active transport, where the transporter utilizes energy, in the form of adenosine triphosphate (ATP) to mobilize molecules against their concentration gradient in order to maintain the correct concentrations of ions and molecules in cells. Symporter transport proteins are co-transporters (such as the glucose–sodium co-transporter): they carry two molecules simultaneously or sequentially in the same direction. Antiporter transport proteins, such as the sodium–potassium pump, carry two molecules simultaneously or sequentially in the opposite direction to each other.
Proteins are generally synthesized on ribosomes in the cytosol or on the rough endoplasmic reticulum: a few are made on mitochondrial ribosomes. Once synthesized, many proteins remain in the cytosol, where they carry out their functions. Others, such as integral membrane proteins or proteins for secretion, are translocated across intracellular membranes for post-translational modification and targeting to their destinations. This is achieved by the signal sequence, an addressing system contained within the protein sequence of amino acids, which is recognized by receptors or translocators in the appropriate membrane. Proteins are sorted by their signal sequence (or set of sequences that become spatially grouped as a signal patch when the protein folds into its tertiary configuration), in order that they are recognized by and enter the correct intracellular membrane compartment.
Plasma membrane proteins also provide surfaces for the attachment of enzymes; sites for the receptors of external signals, including hormones and other ligands; and sites for the recognition and attachment of other cells. Internally, plasma membranes can act as points of attachment for intracellular structures, in particular those concerned with cell motility and other cytoskeletal functions. Cell membranes are synthesized by the rough endoplasmic reticulum in conjunction with the Golgi apparatus.
The external surface of a plasma membrane differs structurally from internal membranes in that it possesses an external, fuzzy, carbohydrate-rich coat, the glycocalyx, composed of the carbohydrate portions of glycoproteins and glycolipids embedded in the plasma membrane (see Fig. 1.3 ). The cell coat forms an integral part of the plasma membrane, projecting as a diffusely filamentous layer 2–20 nm or more from the lipoprotein surface.
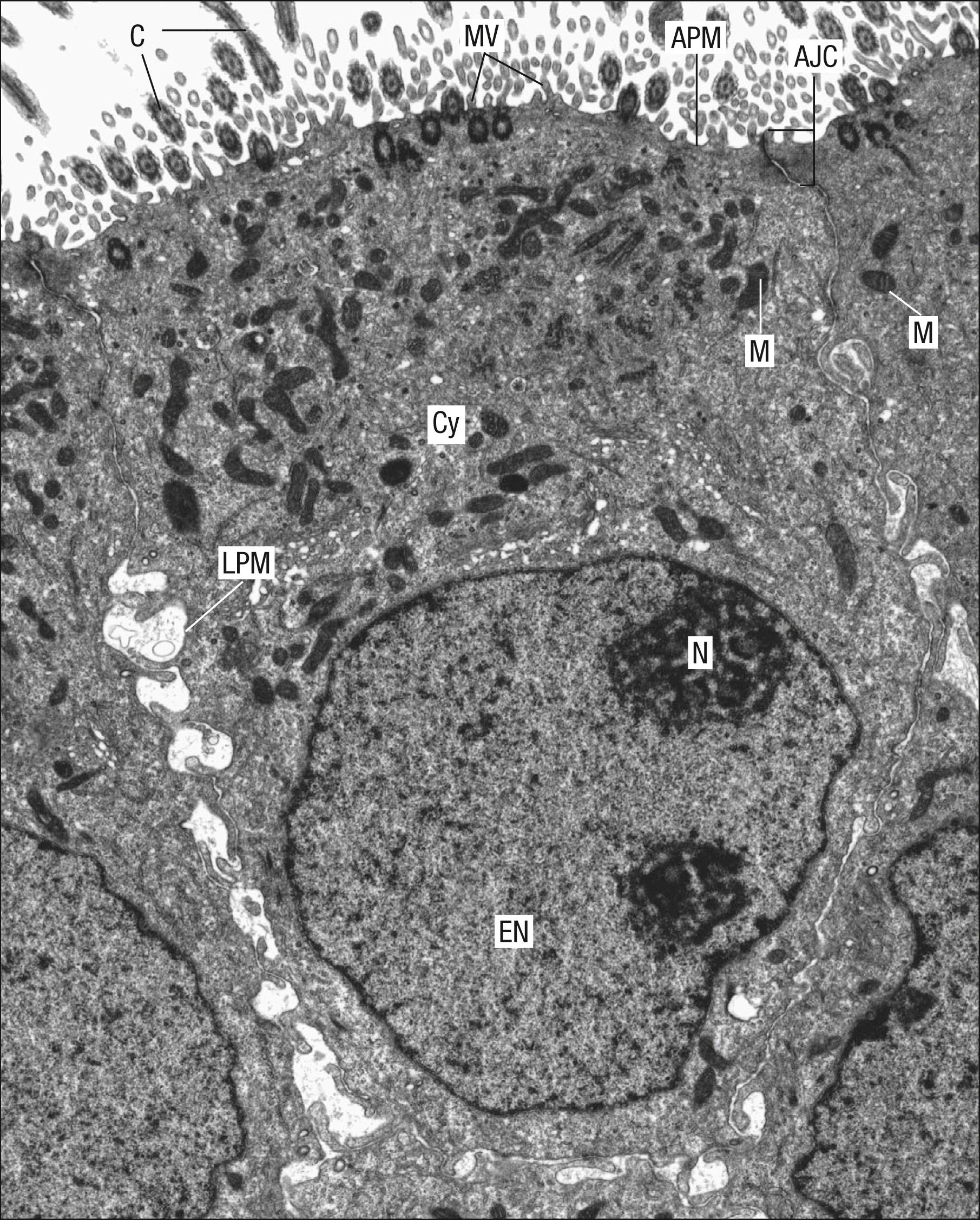
The precise composition of the glycocalyx varies with cell type; many tissue and cell type-specific antigens are located in the coat, including the major histocompatibility complex of the immune system and, in the case of erythrocytes, blood group antigens. The glycocalyx therefore plays a significant role in organ transplant compatibility. The glycocalyx found on the apical microvilli of enterocytes, the cells forming the lining epithelium of the intestine, consists of enzymes involved in the digestive process. Intestinal microvilli are cylindrical projections (1–2 μm long and about 0.1 μm in diameter) forming a closely packed layer called the brush border that increases the absorptive function of enterocytes (see Fig. 1.26 ).
The glycocalyx plays a significant role in maintenance of the integrity of tissues and in a wide range of dynamic cellular processes, e.g. serving as a vascular permeability barrier and transducing fluid shear stress to the endothelial cell cytoskeleton ( ). Disruption of the glycocalyx on the endothelial surface of large blood vessels precedes inflammation, a conditioning factor of atheromatosis (e.g. deposits of cholesterol in the vascular wall leading to partial or complete obstruction of the vascular lumen).
The cytoplasm consists of the cytosol and two cytomembrane systems, the endoplasmic reticulum and Golgi apparatus, all enclosed by the cell or plasma membrane. The cytosol is a gel-like material made up of colloidal proteins such as enzymes, carbohydrates and small protein molecules, together with ribosomes and ribonucleic acids. The cytoplasm also contains membrane-bound organelles (lysosomes, peroxisomes and mitochondria), membrane-free inclusions (glycogen and pigments) and the cytoskeleton. The nuclear contents, the nucleoplasm, are separated from the cytoplasm by the nuclear envelope.
The endoplasmic reticulum is a system of interconnecting membrane-lined channels within the cytoplasm ( Fig. 1.5 ). These channels take various forms, including cisternae (flattened sacs), tubules and vesicles. The membranes divide the cytoplasm into two major compartments. The intramembranous compartment, or cisternal space, is where secretory products are stored or transported to the Golgi complex and cell exterior: it is continuous with the perinuclear space.
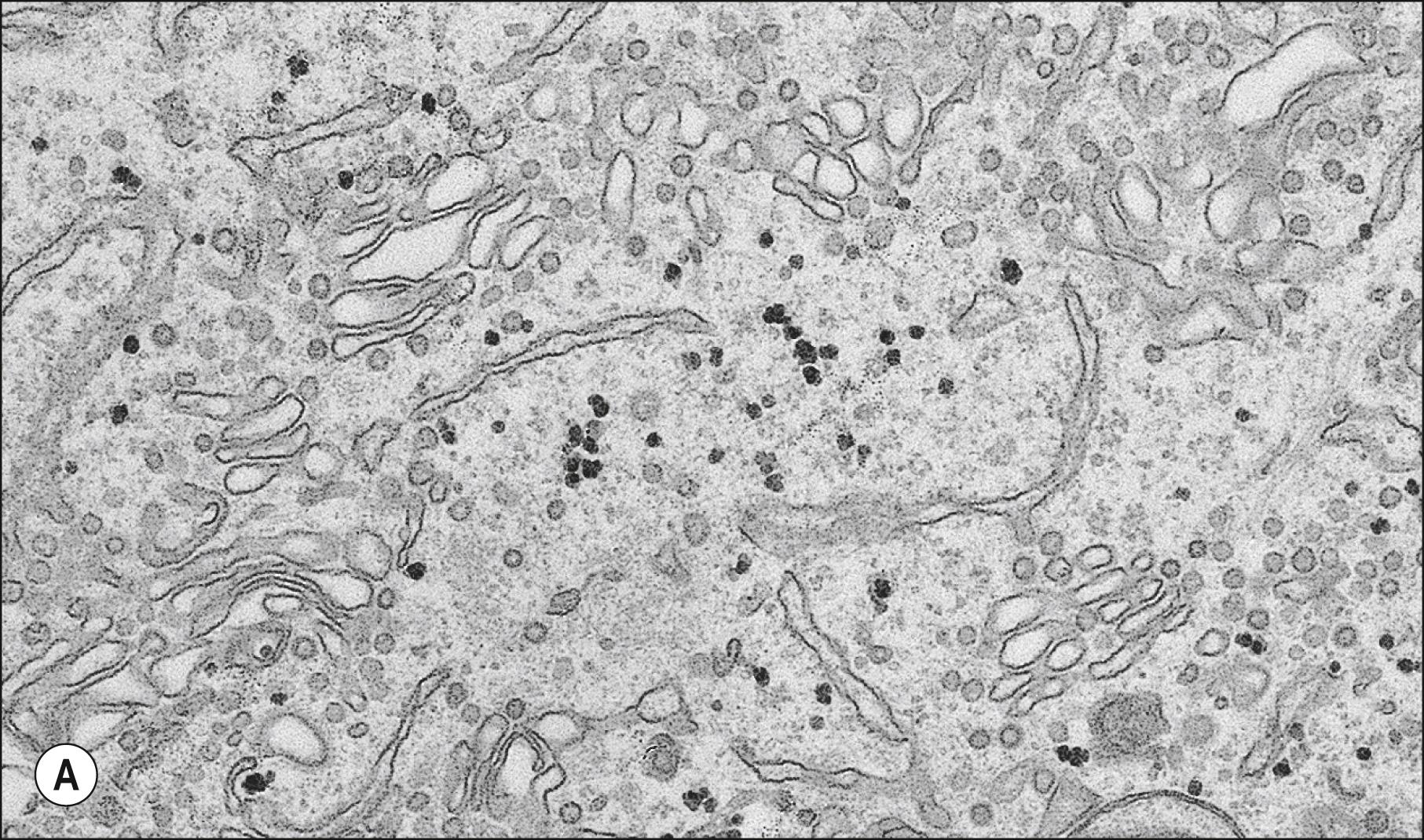
Structurally, the channel system can be divided into rough or granular endoplasmic reticulum (RER), which has ribosomes attached to its outer, cytosolic surface, and smooth or agranular endoplasmic reticulum (SER), which lacks ribosomes. The functions of the endoplasmic reticulum vary greatly and include the synthesis, folding and transport of proteins; the biosynthesis and transport of phospholipids and steroids; the degradation of glycogen; and the maintenance of calcium homeostasis. Endoplasmic reticulum (ER)-associated degradation (ERAD) is part of the cellular protein quality control system and is responsible for the clearance of misfolded proteins from the ER for cytosolic proteasomal degradation.
In general, RER is well developed in cells that produce abundant proteins, whereas SER is abundant in steroid-producing cells and muscle cells. The sarcoplasmic reticulum is a variant of the endoplasmic reticulum in muscle cells: it is involved in calcium storage and release for muscle contraction. For further reading on the endoplasmic reticulum, see .
The smooth endoplasmic reticulum ( Fig. 1.5A ) is associated with carbohydrate metabolism and many other metabolic processes, including detoxification and synthesis of lipids, cholesterol and steroids. The membranes of the smooth endoplasmic reticulum serve as surfaces for the attachment of many enzyme systems, e.g. the enzyme cytochrome P450, which is involved in important detoxification mechanisms and is thus accessible to its substrates, which are generally lipophilic. The smooth endoplasmic reticulum in hepatocytes (which are able to store glycogen) contains the enzyme glucose-6-phosphatase, which converts glucose-6-phosphate to glucose, a step in gluconeogenesis. The membranes also cooperate with the rough endoplasmic reticulum and the Golgi apparatus to synthesize new membranes; the protein, carbohydrate and lipid components are added in different structural compartments.
The rough endoplasmic reticulum is a site of protein synthesis; its cytosolic surface is studded with ribosomes ( Fig. 1.5B ). Ribosomes only bind to the endoplasmic reticulum when proteins targeted for secretion begin to be synthesized. Most proteins pass through its membranes and accumulate within its cisternae, although some integral membrane proteins, e.g. plasma membrane receptors, are inserted into the rough endoplasmic reticulum membrane, where they remain. After passage from the rough endoplasmic reticulum, proteins remain in membrane-bound cytoplasmic organelles such as lysosomes, become incorporated into new plasma membrane, or are secreted by the cell. Some carbohydrates are also synthesized by enzymes within the cavities of the rough endoplasmic reticulum and may be attached to newly formed protein (glycosylation). Vesicles are budded off from the rough endoplasmic reticulum for transport to the Golgi as part of the protein-targeting mechanism of the cell
Ribosomes are macromolecular machines that catalyse the synthesis of proteins from amino acids; synthesis and assembly into subunits takes place in the nucleolus and includes the association of ribosomal RNA (rRNA) with ribosomal proteins translocated from their site of synthesis in the cytoplasm. The individual subunits are then transported into the cytoplasm, where they remain separate from each other when not actively synthesizing proteins. Ribosomes are granules approximately 25 nm in diameter, composed of rRNA molecules and proteins assembled into two unequal subunits. The subunits can be separated by their sedimentation coefficients (S) in an ultracentrifuge into larger 60S and smaller 40S components. These are associated with 73 different proteins (40 in the large subunit and 33 in the small), which have structural and enzymatic functions. Three small, highly convoluted rRNA strands (28S, 5.8S and 5S) make up the large subunit, and one strand (18S) is in the small subunit.
A typical cell contains millions of ribosomes. They may form groups (polyribosomes or polysomes) attached to messenger RNA (mRNA), which they translate during protein synthesis for use outside the system of membrane compartments, e.g. enzymes of the cytosol and cytoskeletal proteins. Some of the cytosolic products include proteins that can be inserted directly into (or through) membranes of selected organelles, such as mitochondria and peroxisomes. Ribosomes may be attached to the membranes of the rough endoplasmic reticulum (see Fig. 1.5B ).
In a mature polyribosome, all the attachment sites of the mRNA are occupied as ribosomes move along it, synthesizing protein according to its nucleotide sequence. Consequently, the number and spacing of ribosomes in a polyribosome indicate the length of the mRNA molecule and hence the size of the protein being made. The two subunits have separate roles in protein synthesis. The 40S subunit is the site of attachment and translation of mRNA. The 60S subunit is responsible for the release of the new protein and, where appropriate, attachment to the endoplasmic reticulum via an intermediate docking protein that directs the newly synthesized protein through the membrane into the cisternal space.
Protein synthesis on ribosomes may be suppressed by a class of RNA molecules known as small interfering RNA (siRNA) or silencing RNA. These molecules are typically 20–25 nucleotides in length and bind (as a complex with proteins) to specific mRNA molecules via their complementary sequence. This triggers the enzymatic destruction of the mRNA or prevents the movement of ribosomes along it. Synthesis of the encoded protein is thus prevented. Their normal function may have antiviral or other protective effects; there is also potential for developing artificial siRNAs as a therapeutic tool for silencing disease-related genes.
The Golgi apparatus is a distinct cytomembrane system located near the nucleus and the centrosome. It is particularly prominent in secretory cells and can be visualized when stained with silver or other metallic salts. Traffic between the endoplasmic reticulum and the Golgi apparatus is bidirectional and takes place via carrier vesicles derived from the donor site that bud, tether and fuse with the target site. Carrier vesicles in transit from the endoplasmic reticulum to the Golgi apparatus (anterograde transport) are coated by coat protein complex II (COPII), whereas COPI-containing vesicles function in the retrograde transport route from the Golgi apparatus (reviewed in ).
Golgins are long coiled-coil proteins attached to the cytoplasmic surface of cisternal membranes, forming a fibrillar matrix surrounding the Golgi apparatus to stabilize it; they have a role in vesicle trafficking ( ). The Golgi apparatus has several functions: it links anterograde and retrograde protein and lipid flow in the secretory pathway; it is the site where protein and lipid glycosylation occurs; and it provides membrane platforms to which signalling and sorting proteins bind.
Ultrastructurally, the Golgi apparatus ( Fig. 1.6A ) displays a continuous ribbon-like structure consisting of a stack of several flattened membranous cisternae, together with clusters of vesicles surrounding its surfaces. Cisternae differ in their enzymatic content and activity. Small transport vesicles from the rough endoplasmic reticulum are received at one face of the Golgi stack, the convex cis -face (entry or forming surface). Here, they deliver their contents to the first cisterna in the series by membrane fusion. The protein is transported from the edges of this cisterna to the next cisterna by vesicular budding and then fusion, and this process is repeated across medial cisternae until the final cisterna at the concave trans -face (exit or condensing surface) is reached. Here, larger vesicles are formed for delivery to other parts of the cell.
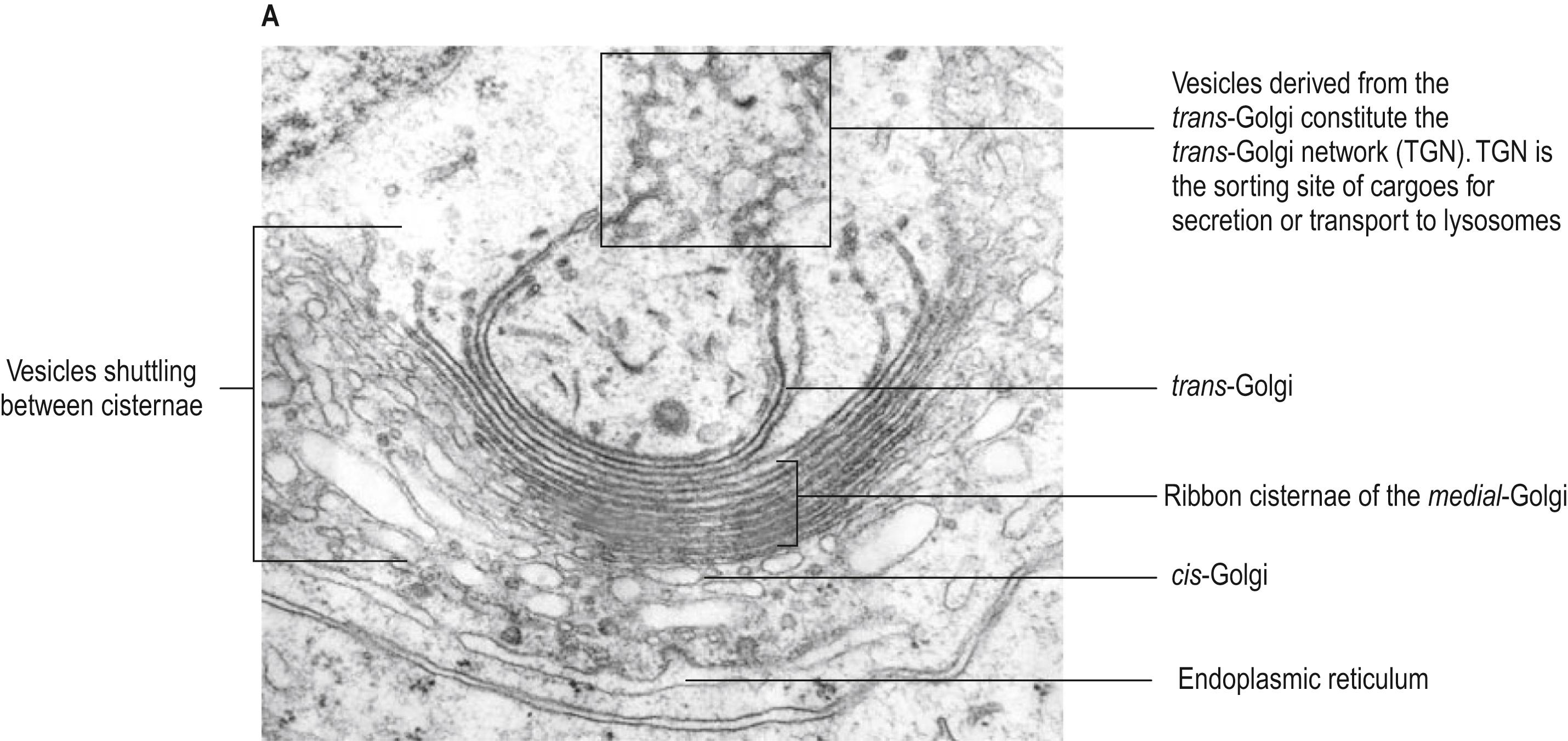
The cis -Golgi and trans -Golgi membranous networks form an integral part of the Golgi apparatus ( Fig. 1.6B ). The cis -Golgi network is a region of complex membranous channels interposed between the rough endoplasmic reticulum and the Golgi cis -face, which receives and transmits vesicles in both directions. Its function is to select appropriate proteins synthesized on the rough endoplasmic reticulum for delivery by vesicles to the Golgi stack, while inappropriate proteins are shuttled back to the rough endoplasmic reticulum.
The trans -Golgi network, at the other side of the Golgi stack, is also a region of interconnected membrane channels engaged in protein sorting. Here, modified proteins processed in the Golgi cisternae are packaged selectively into vesicles and dispatched to different parts of the cell. The packaging depends on the detection, by the trans -Golgi network, of particular amino acid signal sequences, leading to their enclosure in membranes of appropriate composition that will further modify their contents, e.g. by extracting water to concentrate them (vesicles entering the exocytosis pathway) or by pumping in protons to acidify their contents (lysosomes destined for the intracellular sorting pathway). The membranes contain specific signal proteins that may allocate them to microtubule-based transport pathways and allow them to dock with appropriate targets elsewhere in the cell, e.g. the plasma membrane in the case of secretory vesicles. Vesicle formation and budding at the trans -Golgi network involves the addition of clathrin on their external surface, to form coated pits.
Within the Golgi stack proper, proteins undergo a series of sequential chemical modifications by Golgi resident enzymes synthesized in the rough endoplasmic reticulum. These include glycosylation (changes in glycosyl groups, e.g. removal of mannose, addition of N -acetylglucosamine and sialic acid); sulphation (addition of sulphate groups to glycosaminoglycans); and phosphorylation (addition of phosphate groups). Some modifications serve as signals to direct proteins and lipids to their final destination within cells, including lysosomes and plasma membrane. Lipids formed in the endoplasmic reticulum are also routed for incorporation into vesicles.
Secreted proteins, lipids, glycoproteins, small molecules such as amines, and other cellular products destined for export from the cell are transported to the plasma membrane in small vesicles released from the trans -face of the Golgi apparatus. This pathway is either constitutive, in which transport and secretion occur more or less continuously, as with immunoglobulins produced by plasma cells, or it is regulated by external signals, as in the control of salivary secretion by autonomic neural stimulation. In regulated secretion, the secretory product is stored temporarily in membrane-bound secretory granules or vesicles. Exocytosis is achieved by fusion of the secretory vesicular membrane with the plasma membrane and release of the vesicle contents into the extracellular domain. In polarized cells, e.g. most epithelia, exocytosis occurs at the apical plasma membrane. Glandular epithelial cells secrete into a duct lumen, as in the pancreas, or on to a free surface, such as the lining of the stomach. In hepatocytes, bile is secreted across a very small area of plasma membrane forming the wall of the bile canaliculus. This region, the apical plasma membrane, is the site of exocrine secretion, whereas secretion of hepatocyte plasma proteins into the blood stream is targeted to the basolateral surfaces facing the sinusoids. Packaging of different secretory products into appropriate vesicles takes place in the trans -Golgi network. Delivery of secretory vesicles to their correct plasma membrane domains is achieved by sorting sequences in the cytoplasmic tails of vesicular membrane proteins.
The endocytic pathway begins at the plasma membrane and ends in lysosomes (see below) involved in the degradation of the endocytic cargo through the enzymatic activity of lysosomal hydrolases. Endocytic cargo is internalized from the plasma membrane to early endosomes and then to late endosomes. Late endosomes transport their cargo to lysosomes, where the cargo material is degraded following fusion and mixing of contents of endosomes and lysosomes. Early endosomes derive from endocytic vesicles (clathrin-coated vesicles and caveolae). Once internalized, endocytic vesicles shed their coat of adaptin and clathrin, and fuse to form an early endosome, where the receptor molecules release their bound ligands. Membrane and receptors from the early endosomes can be recycled to the cell surface as exocytic vesicles.
Clathrin-dependent endocytosis occurs at specialized patches of plasma membrane called coated pits: this mechanism is also used to internalize ligands bound to surface receptor molecules and is also termed receptor-mediated endocytosis. Caveolae are structurally distinct pinocytotic vesicles most widely used by endothelial and smooth muscle cells, where they are involved in transcytosis, signal transduction and possibly other functions. In addition to late endosomes, lysosomes can also fuse with phagosomes, autophagosomes and plasma membrane patches for membrane repair. Lysosomal hydrolases process or degrade exogenous materials (phagocytosis or heterophagy) as well as endogenous material (autophagy). Phagocytosis consists of the cellular uptake of invading pathogens, apoptotic cells and other foreign material by specialized cells. Lysosomes are numerous in actively phagocytic cells, e.g. macrophages and neutrophil granulocytes, where they are responsible for destroying phagocytosed particles, e.g. bacteria. These cells may contain phagosomes, vesicles that potentially contain a pathogenic microorganism and that may fuse with several lysosomes.
Specialized cells of the immune system, antigen-presenting cells (APCs), degrade protein molecules (antigens) transported by the endocytic pathway for lysosomal breakdown, and expose their fragments to the cell exterior to elicit an immune response, that is mediated initially by helper T cells.
Late endosomes receive lysosomal enzymes from primary lysosomes ( Fig. 1.6C ) derived from the Golgi apparatus after late endosome–lysosome membrane tethering and fusion followed by diffusion of lysosomal contents into the endosomal lumen. The pH inside the fused hybrid organelle, now called a secondary lysosome, is low (about 5.0): this activates lysosomal acid hydrolases to degrade the endosomal contents. The products of hydrolysis are either passed through the membrane into the cytosol, or they may be retained in the secondary lysosome ( Fig. 1.6D ). Secondary lysosomes may grow considerably in size by vesicle fusion to form multivesicular bodies; the enzyme concentration may increase greatly to form large lysosomes.
Lysosomes are membrane-bound organelles 80–800 nm in diameter, often with complex inclusions of material undergoing hydrolysis (secondary lysosomes). Two classes of protein participate in lysosomal function, namely soluble acid hydrolases and integral lysosomal membrane proteins. Each of the 60 or more known acid hydrolases (including proteases, lipases, carbohydrases, esterases and nucleases) degrades a specific substrate. There are about 25 lysosomal membrane proteins participating in the acidification of the lysosomal lumen, protein import from the cytosol, membrane fusion and transport of degradation products to the cytoplasm. Material that has been hydrolysed within secondary lysosomes may be completely degraded to soluble products, e.g. amino acids, which are recycled through metabolic pathways. However, degradation is usually incomplete and some debris remains. A debris-laden vesicle is called a residual body or tertiary lysosome (see Fig. 1.6B ), and may be passed to the cell surface, where it is ejected by exocytosis; alternatively, it may persist inside the cell as an inert residual body. Considerable numbers of residual bodies can accumulate in long-lived cells, often fusing to form larger dense vacuoles with complex lamellar inclusions. As their contents are often darkly pigmented, this may change the colour of the tissue; e.g. in neurones, the end-product of lysosomal digestion, lipofuscin (neuromelanin or senility pigment), gives ageing brains a brownish-yellow coloration. Lysosomal enzymes may also be secreted – often as part of a process to alter the extracellular matrix, as in osteoclast-mediated erosion during bone resorption. For further reading on lysosome biogenesis, see .
The transcription factor EB (TFEB) is responsible for regulating lysosomal biogenesis and function, lysosome-to-nucleus signalling and lipid catabolism (for further reading, see ). If there is a failure of any of the actions of lysosomal hydrolases, of the lysosome acidification mechanism or of lysosomal membrane proteins, there is progressive lysosomal dysfunction in several tissues and organs. This may be due to faulty degradation and recycling of extracellular substrates delivered to lysosomes by the late endosome or the defective degradation and recycling of intracellular substrates by autophagy.
Lysosomal storage disorders (LSDs) are a consequence of lysosomal dysfunction. Approximately 60 different types of LSD have been identified on the basis of the type of material accumulated in cells (such as mucopolysaccharides, sphingolipids, glycoproteins, glycogen and lipofuscins).
LSDs are characterized by severe neurodegeneration, mental decline, and cognitive and behavioural abnormalities. Autophagy impairment caused by defective lysosome–autophagosome coupling triggers a pathogenic cascade by the accumulation of substrates that contribute to neurodegenerative disorders, including Parkinson's disease, Alzheimer's disease, Huntington's disease and several tau-opathies. In Tay–Sachs disease (GM2 gangliosidosis), a faulty β-hexosaminidase A leads to the accumulation of the glycosphingolipid GM2 ganglioside in neurones, causing death during childhood. In Danon disease, a vacuolar skeletal myopathy and cardiomyopathy with neurodegeneration in hemizygous males, lysosomes fail to fuse with autophagosomes because of a mutation of the lysosomal membrane protein LAMP-2 (lysosomal-associated membrane protein-2).
Peroxisomes are small (0.2–1 μm in diameter) membrane-bound organelles present in most mammalian cells. They contain more than 50 enzymes responsible for multiple catabolic and synthetic biochemical pathways, in particular the β-oxidation of very-long-chain fatty acids (>C22) and the metabolism of hydrogen peroxide (hence the name peroxisome). Peroxisomes may be derived from pre-existing peroxisomes that divide by fission. Alternatively, they may arise de novo by budding as pre-peroxisomal vesicles from the endoplasmic reticulum and possibly from mitochondria ( ). All matrix proteins and some peroxisomal membrane proteins are synthesized by cytosolic ribosomes and contain a peroxisome targeting signal that enables them to be imported by proteins called peroxins ( , ).
Peroxisomes often contain crystalline inclusions composed mainly of high concentrations of the enzyme urate oxidase. Oxidases use molecular oxygen to oxidize specific organic substrates (such as l -amino acids, d -amino acids, urate, xanthine and very-long-chain fatty acids) and produce hydrogen peroxide that is detoxified (degraded) by peroxisomal catalase. Peroxisomes are important in the oxidative detoxification of various substances taken into or produced within cells. They are particularly numerous in hepatocytes.
Peroxin mutation is a characteristic feature of Zellweger syndrome (craniofacial dysmorphism and malformations of brain, liver, eye and kidney; cerebrohepatorenal syndrome). Neonatal leukodystrophy is an X-linked peroxisomal disease affecting mostly males, caused by deficiency in β-oxidation of very-long-chain fatty acids.
The build-up of very-long-chain fatty acids in the nervous system and suprarenal glands determines progressive deterioration of brain function and suprarenal insufficiency (Addison's disease). For further reading, see .
Mitochondria appear in the light microscope as long, thin structures in the cytoplasm of most cells, particularly those with a high metabolic rate, e.g. secretory cells in exocrine glands. In the electron microscope, mitochondria usually appear as round or elliptical bodies 0.5–2.0 μm long ( Fig. 1.7 ), consisting of an outer mitochondrial membrane; an inner mitochondrial membrane, separated from the outer membrane by an intermembrane space; cristae, infoldings of the inner membrane that harbour ATP synthase to generate ATP; and the mitochondrial matrix, a space enclosed by the inner membrane and numerous cristae. The permeability of the two mitochondrial membranes differs considerably: the outer membrane is freely permeable to many substances because of the presence of large non-specific channels formed by proteins (porins), whereas the inner membrane is permeable to only a narrow range of molecules. The presence of cardiolipin, a phospholipid, in the inner membrane may contribute to this relative impermeability.
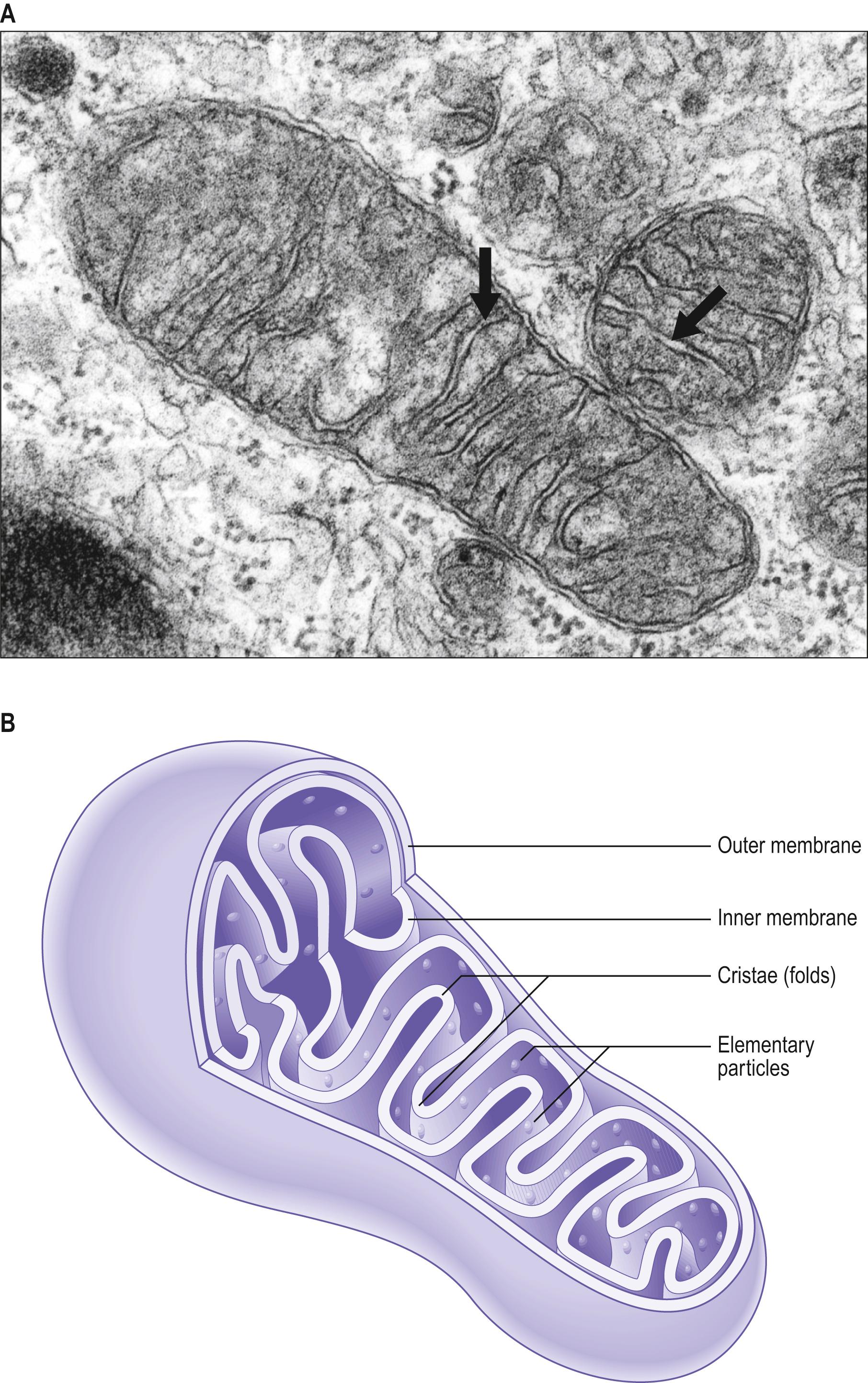
Mitochondria are the principal source of chemical energy in most cells. They are the site of the citric acid (Krebs') cycle and the electron transport (cytochrome) pathway by which complex organic molecules are finally oxidized to carbon dioxide and water. This process provides the energy to drive the production of ATP from adenosine diphosphate (ADP) and inorganic phosphate (oxidative phosphorylation). The various enzymes of the citric acid cycle are located in the mitochondrial matrix, whereas those of the cytochrome system and oxidative phosphorylation are localized chiefly in the inner mitochondrial membrane. The intermembrane space houses cytochrome c , a molecule involved in activation of apoptosis.
Cristae are abundant in mitochondria within cardiac muscle cells and in steroid-producing cells (in the suprarenal cortex, corpus luteum and Leydig cells). The protein, steroidogenic acute regulatory protein (StAR), regulates the synthesis of steroids by transporting cholesterol across the outer mitochondrial membrane. A mutation in the gene encoding StAR causes defective suprarenal and gonadal steroidogenesis.
The number of mitochondria in a particular cell reflects its general energy requirements; e.g. in hepatocytes there may be as many as 2000, whereas in resting lymphocytes there are usually very few. Mature erythrocytes lack mitochondria altogether. Cells with few mitochondria generally rely largely on glycolysis for their energy supplies. These include some very active cells, e.g. fast-twitch skeletal muscle fibres, which are able to work rapidly but for only a limited duration. Mitochondria are distributed within a cell according to regional energy requirements, e.g. near the bases of cilia in ciliated epithelia, in the basal domain of the cells of proximal convoluted tubules in the renal cortex (where considerable active transport occurs) and around the proximal segment, called the middle piece, of the flagellum in spermatozoa. In living cells, mitochondria constantly change shape and intracellular position; they multiply by growth and fission, and may undergo fusion.
Mitochondria may be involved in tissue-specific metabolic reactions, e.g. various urea-forming enzymes are found in liver cell mitochondria. Moreover, a number of genetic diseases of mitochondria affect particular tissues exclusively, e.g. mitochondrial myopathies (skeletal muscle) and mitochondrial neuropathies (nervous tissue).
The mitochondrial matrix is an aqueous environment. It contains a variety of enzymes, and strands of mitochondrial DNA with the capacity for transcription and translation of a unique set of mitochondrial genes (mitochondrial mRNAs and transfer RNAs, mitochondrial ribosomes with rRNAs). The DNA forms a closed loop, about 5 μm across and several identical copies are present in each mitochondrion. The ratio between its bases differs from that of nuclear DNA; the RNA sequences also differ in the precise genetic code used in protein synthesis. At least 13 respiratory chain enzymes of the matrix and inner membrane are encoded by the small number of genes along the mitochondrial DNA (mtDNA). The great majority of mitochondrial proteins are encoded by nuclear genes and made in the cytosol, then inserted through special channels in the mitochondrial membranes to reach their destinations. Their membrane lipids are synthesized in the endoplasmic reticulum.
Mitochondrial ribosomes are smaller and quite distinct from those of the rest of the cell in that they (and mitochondrial nucleic acids) resemble those of bacteria. This similarity underpins the theory that mitochondrial ancestors were oxygen-utilizing bacteria that existed in a symbiotic relationship with eukaryotic cells unable to metabolize the oxygen produced by early plants.
It has been shown that mitochondria are of maternal origin because the mitochondria of spermatozoa are not generally incorporated into the ovum at fertilization. As mitochondria are formed only from previously existing ones, it follows that all mitochondria in the body are descended from those in the cytoplasm of the ovum. Thus mitochondria (and mitochondrial genetic variations and mutations) are passed only through the female line. For further information on mitochondrial genetics and disorders, see .
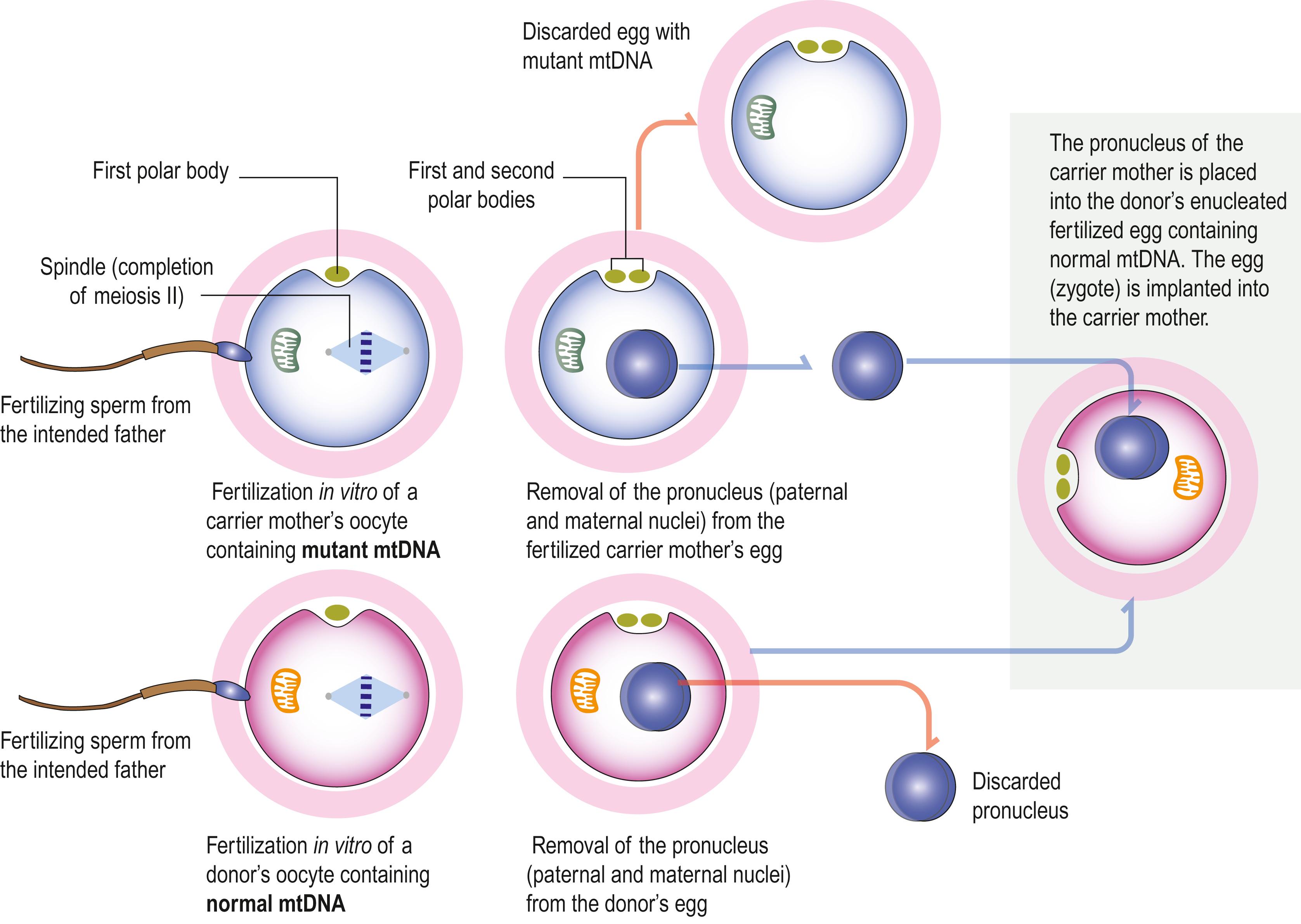
Mitochondrial replacement therapies have been developed to resolve mitochondrial disorders of maternal origin, e.g. by transplantating the pronucleus from an egg containing mutant mtDNA from a carrier mother to an enucleated egg from an unaffected donor ( Fig. 1.8 ). This procedure can be performed after in vitro fertilization using sperm from the intended father. Soon after fertilization, the haploid maternal and paternal nuclei are assembled as a pronucleus. Because it is difficult to distinguish between male and female pronuclei, pronuclear transfer (PNT) involves transplantation of both the maternal and paternal pronuclei, clearly visualized by light microscopy. mtDNA resides within mitochondria, separately from the genes housed in each cell nucleus. Since mitochondria are inherited only through the maternal germline, mutated mitochondria are thus replaced with normal mitochondria from enucleated oocytes provided by the unaffected donor. An alternative procedure involves transfer of the nuclear genetic material from the affected female (maternal spindle transfer, MST) into an enucleated donor egg with normal mitochondria, prior to fertilization.
The aqueous cytosol surrounds the membranous organelles described above. It also contains various non-membranous inclusions, including free ribosomes, components of the cytoskeleton, and other inclusions, such as storage granules (e.g. glycogen), pigments (such as lipofuscin granules, remnants of the lipid oxidative mechanism seen in the suprarenal cortex).
Lipid droplets are spherical bodies of various sizes found within many cells, but are especially prominent in the adipocytes (fat cells) of adipose connective tissue. They do not belong to the Golgi-related vacuolar system of the cell. Lipid droplets, suspended in the cytosol, are surrounded by perilipin proteins, which regulate lipid storage and lipolysis. (See for further reading on obesity and perilipins.) In cells specialized for lipid storage, the vacuoles reach 80 μm or more in diameter. They function as stores of chemical energy, thermal insulators and mechanical shock absorbers in adipocytes. In many cells, they may represent end-products of other metabolic pathways, e.g. in steroid-synthesizing cells, where they are a prominent feature of the cytoplasm. They may also be secreted, as in the alveolar epithelium of the lactating breast.
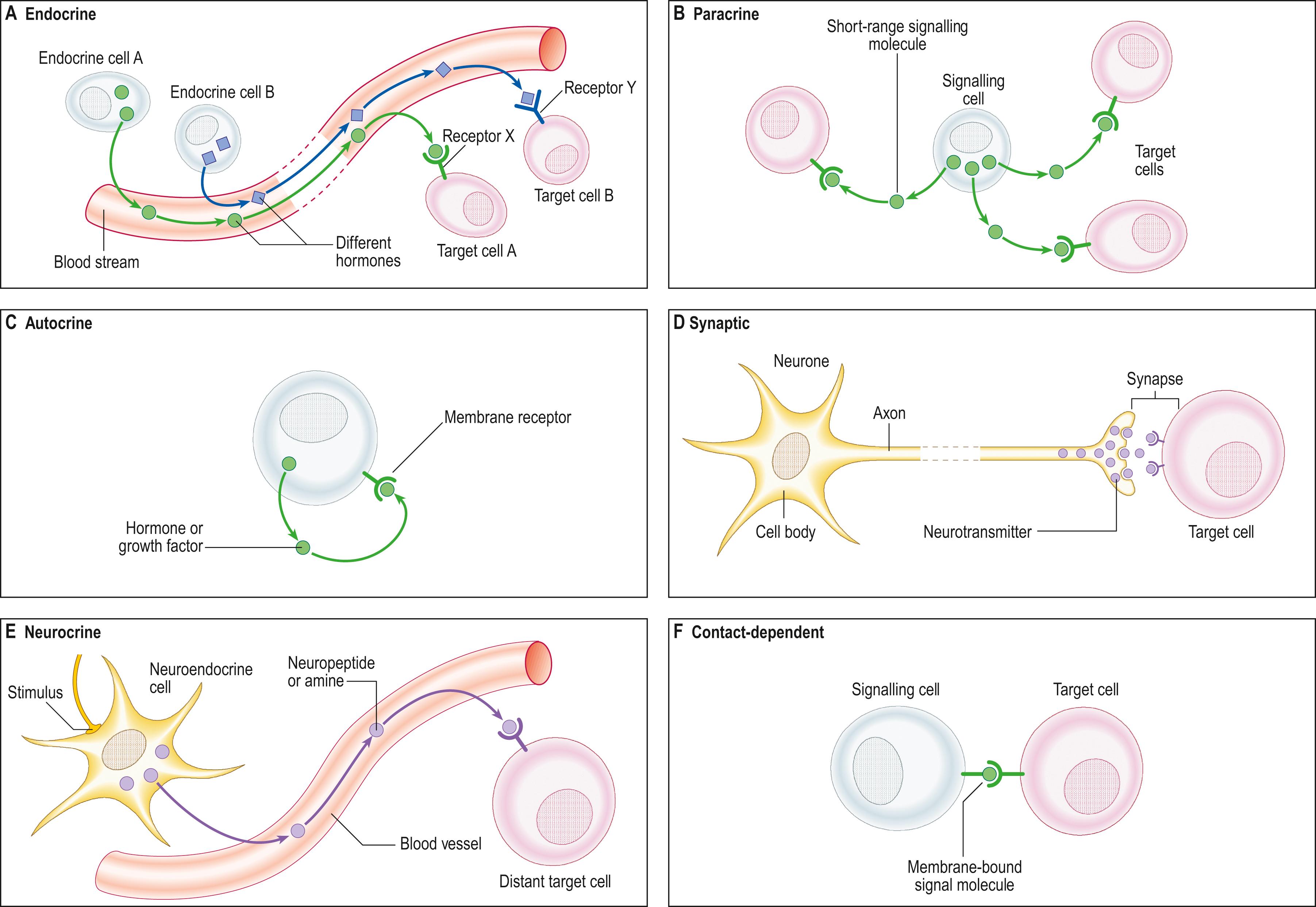
Cellular systems in the body communicate with each other to coordinate and integrate their functions, through a variety of processes known collectively as cell signalling, in which a signalling molecule produced by one cell is detected by another, almost always by means of a specific receptor protein molecule. The recipient cell transduces the signal, which it most often detects at the plasma membrane, into intracellular chemical messages that change cell behaviour.
The signal may act over a long distance, e.g. endocrine signalling through the release of hormones into the blood stream, or neuronal synaptic signalling via electrical impulse transmission along axons and subsequent release of chemical transmitters of the signal at synapses or neuromuscular junctions. A specialized variation of endocrine signalling (neurocrine or neuroendocrine signalling) occurs when neurones or paraneurones (e.g. chromaffin cells of the suprarenal medulla) secrete a hormone into interstitial fluid and the blood stream. Alternatively, signalling may occur at short range through a paracrine mechanism, in which cells of one type release molecules into the interstitial fluid of the local environment, to be detected by nearby cells of a different type that express the specific receptor protein.
Neurocrine cell signalling uses chemical messengers found also in the central nervous system, which may act in a paracrine manner via interstitial fluid or reach more distant target tissues via the blood stream. Cells may generate and respond to the same signal. This is autocrine signalling, a phenomenon that reinforces the coordinated activities of a group of like cells, which respond together to a high concentration of a local signalling molecule. The most extreme form of short-distance signalling is contact-dependent (juxtacrine) signalling, where one cell responds to transmembrane proteins of an adjacent cell that bind to surface receptors in the responding cell membrane. Contact-dependent signalling also includes cellular responses to integrins on the cell surface binding to elements of the extracellular matrix. Juxtacrine signalling is important during development and in immune responses. These different types of intercellular signalling mechanism are illustrated in Fig. 1.9 .
The majority of signalling molecules (ligands) are hydrophilic and so cannot cross the plasma membrane of a recipient cell to effect changes inside the cell unless they first bind to a plasma membrane receptor protein. Ligands are mainly proteins (usually glycoproteins), polypeptides or highly charged biogenic amines. They include classic peptide hormones of the endocrine system; cytokines, which are mainly of haemopoietic cell origin and involved in inflammatory responses and tissue remodelling (e.g. the interferons, interleukins, tumour necrosis factor, leukaemia inhibitory factor); and polypeptide growth factors (e.g. the epidermal growth factor superfamily, nerve growth factor, platelet-derived growth factor, the fibroblast growth factor family, transforming growth factor beta and the insulin-like growth factors). Polypeptide growth factors are multifunctional molecules with more widespread actions and cellular sources than their names suggest. They and their receptors are commonly mutated or aberrantly expressed in certain cancers. The cancer-causing gene variant is termed a transforming oncogene and the normal (wild-type) version of the gene is a cellular oncogene or proto-oncogene. The activated receptor acts as a transducer to generate intracellular signals, which are either small diffusible second messengers (e.g. calcium, cyclic adenosine monophosphate or the plasma membrane lipid-soluble diacylglycerol), or larger protein complexes that amplify and relay the signal to target control systems.
Some signals are hydrophobic and able to cross the plasma membrane freely. Classic examples are the steroid hormones, thyroid hormones, retinoids and vitamin D. Steroids, for instance, enter cells non-selectively, but elicit a specific response only in those target cells that express specific cytoplasmic or nuclear receptors. Light stimuli also cross the plasma membranes of photoreceptor cells and interact intracellularly, at least in rod cells, with membrane-bound photosensitive receptor proteins. Hydrophobic ligands are transported in the blood stream or interstitial fluids, generally bound to carrier proteins, and they often have a longer half-life and longer-lasting effects on their targets than do water-soluble ligands.
A separate group of signalling molecules that are able to cross the plasma membrane freely is typified by the gas, nitric oxide. The principal target of short-range nitric oxide signalling is smooth muscle, which relaxes in response. Nitric oxide is released from vascular endothelium as a result of the action of autonomic nerves that supply the vessel wall, causing local relaxation of smooth muscle and consequent dilation of the vessel. This mechanism is responsible for penile erection. Nitric oxide is unusual among signalling molecules in having no specific receptor protein; it acts directly on intracellular enzymes of the response pathway.
There are some 20 different families of receptor proteins, each with several isoforms responding to different ligands. The great majority of these receptors are transmembrane proteins. Members of each family share structural features that indicate either shared ligand-binding characteristics in the extracellular domain or shared signal transduction properties in the cytoplasmic domain, or both. There is little relationship either between the nature of a ligand and the family of receptor proteins to which it binds and activates, or the signal transduction strategies by which an intracellular response is achieved. The same ligand may activate fundamentally different types of receptor in different cell types.
Cell surface receptor proteins are generally grouped according to their linkage to one of three intracellular systems: ion channel-linked receptors; G-protein coupled receptors; or receptors that link to enzyme systems. Other receptors do not fit neatly into any of these categories. All the known G-protein-coupled receptors belong to a structural group of proteins that pass through the membrane seven times in a series of serpentine loops: these receptors are thus known as seven-pass transmembrane receptors or, because the transmembrane regions are formed from α-helical domains, as seven-helix receptors. The best known of this large group of phylogenetically ancient receptors are the odorant-binding proteins of the olfactory system; the light-sensitive receptor protein, rhodopsin; and many of the receptors for clinically useful drugs. A comprehensive list of receptor proteins, their activating ligands and examples of the resultant biological function is given in .
Most cell surface receptors stimulate intracellular target enzymes to transmit and amplify a signal upon ligand binding. A signalling pathway can activate a series of intracellular targets located downstream of the receptor, in particular the activity of intracellular proteins, or, like neurotransmitter receptors, control the flow of water (aquaporins) and electrolytes across ligand-gated ion channels located on the plasma membrane. An amplified signal can be propagated to the nucleus to regulate gene expression in response to an external cell stimulus.
The cyclic adenosine monophosphate (cAMP) pathway is a major intracellular signalling pathway. For example, there is an increase in the intracellular concentration of cAMP when adrenaline (epinephrine) binds to its receptor. Adrenaline breaks down glycogen into glucose before muscle contraction. cAMP is formed from ATP by the action of the enzyme adenylyl cyclase and degraded to adenosine monophosphate (AMP) by the enzyme cAMP phosphodiesterase. This mechanism led to the concept of a first messenger (adrenaline) mediating a cell-signalling effect by a second messenger, cAMP.
The adrenaline receptor is linked to adenylyl cyclase by G protein, which stimulates cyclase activity upon adrenaline binding. In the adrenaline-dependent regulation of glycogen metabolism, the intracellular signalling effects of cAMP are mediated by the enzyme cAMP-dependent protein kinase (or protein kinase A). In its inactive form, protein kinase A is a tetramer composed of two regulatory subunits (to which cAMP binds) and two catalytic subunits. Binding of cAMP results in the dissociation of the catalytic subunits. Free catalytic subunits can phosphorylate serine residues on target proteins ( Fig. 1.10 ).

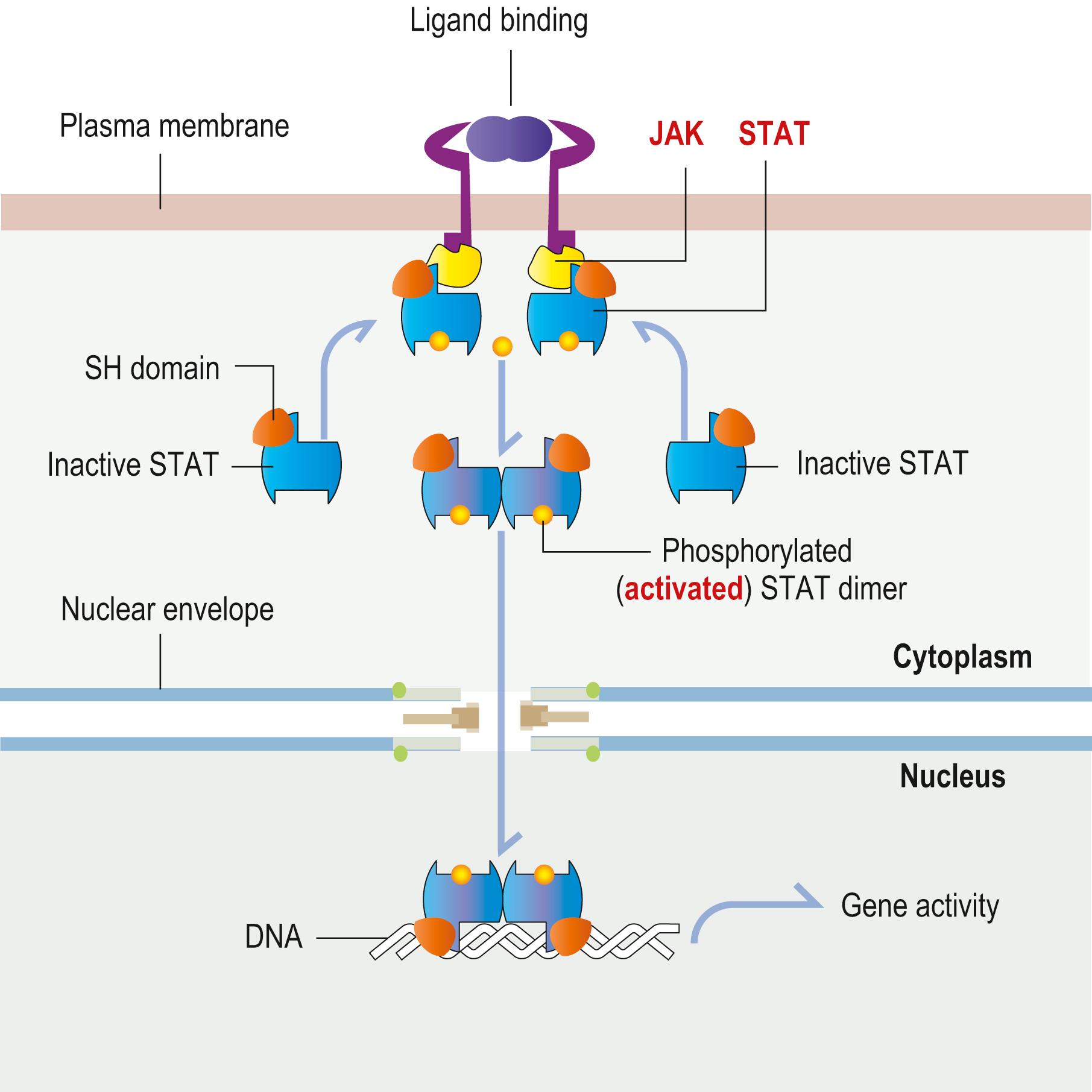
Signalling pathways play important roles in embryonic and fetal development, body axis patterning, cell migration and cell proliferation. They all contain components subject to diverse regulatory steps and crosstalk mechanisms and some use different downstream effectors activated by specific transcription factors. The following signalling pathways have clinical relevance: the JAK–STAT, Hedgehog (Hh), Wingless (Wnt) and transforming growth factor (TGF)-β signalling pathways. For example, erythropoietin stimulates the development of the erythroid lineage (red blood cell formation) in bone marrow by a mechanism that involves the JAK–STAT pathway ( Fig. 1.11 ).
This pathway links a protein with tyrosine kinase activity (JAK) to a transcription factor (STAT) that becomes activated by this event. STAT (Signal Transducers and Activators of Transcription) proteins are transcription factors with an SH2 domain. They are present in the cytoplasm in an inactive state. Stimulation of a receptor by ligand binding recruits STAT proteins, which bind to the cytoplasmic portion of the receptor-associated JAK protein tyrosine kinase through their SH2 domain and become phosphorylated. Phosphorylated STAT proteins then dimerize and translocate into the nucleus, where they activate the transcription of target genes ( Fig. 1.11 ).
The Hh signalling pathway ( Fig. 1.12A ) is involved in switching Gli (Glioma) factors from transcriptional repressors into activators in the cytoplasm to allow Hh-specific transcriptional events. Hh proteins bind to the receptor PTCH1 (PaTCHed Homologue 1) and then signal through SMO (SMOothened), a transmembrane protein, to regulate gene transcription by repressing or activating Gli3, the transcription factor. If SMO is not present, Sufu (SUppressor of FUsed) permits the truncated Gli3 repressor to block Hh-specific gene expression. If SMO is present, activated full-length Gli2A translocates to the cell nucleus to regulate Hh-specific gene expression (the expression of cyclin D, cyclin E, Myc and Patched). Hh ligands include Sonic (Shh), Indian (Ihh) and Desert (Dhh). Dysfunctions of the Hh signalling pathway include Gorlin syndrome, basal cell carcinoma in the skin and medulloblastoma.
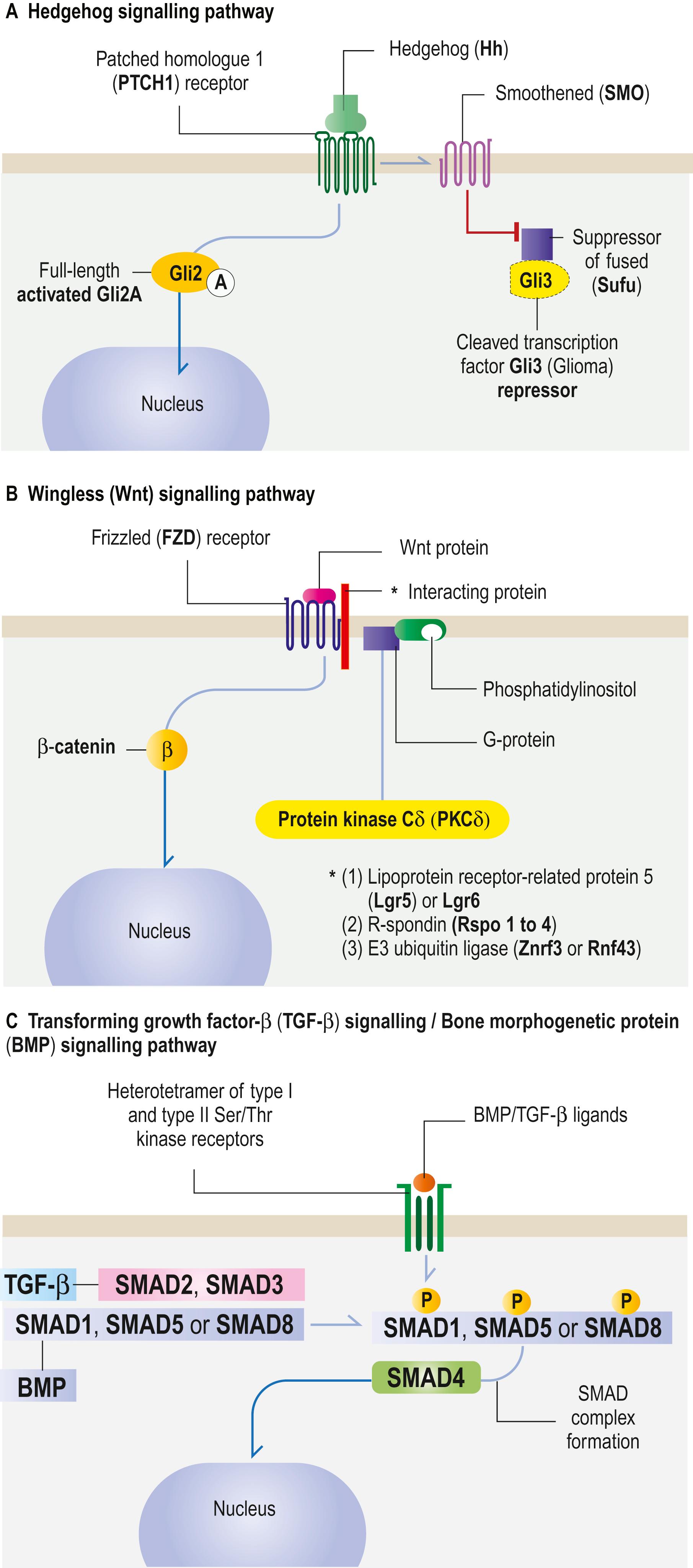
The Wingless (Wnt) signalling pathway ( Fig. 1.12B ) relies on Wnt proteins controlling tissue organization and body-axis formation during embryogenesis and the regulation of stem cells in adult tissue. Wnt protein binding to FZD (frizzled) receptor is regulated by several interacting proteins, including leucine-rich repeat containing G protein-coupled receptor (Lgr) 5 or 6; R-spondin (Rspo) 1 to 4 and E3 ubiquitin ligase enzyme (Znrf3 or Rnf43). In the absence of Rspos, the ubiquitin ligase catalyses FZD degradation, thus preventing Wnt protein binding. Rspo prevents enzyme activity, FZD receptors accumulate and Wnt signalling increases. β-catenin then translocates to the nucleus and stimulates the transcription of Wnt target genes by interacting with the co-activators LEF1 (Lymphoid Enhancer-binding Factor 1) and TCF (T Cell Factor) 1, TCF3 and TCF4 (not shown in Fig. 1.12B ). In the β-catenin independent pathway, Wnt protein induces G protein-coupled phosphatidylinositol to activate protein kinase C (PKC). Wnt signalling has direct implications for regenerative medicine and Wnt-associated cancers. Regeneration, in contrast to repair, is a steady-state condition, for example of the normal epidermis and the gastrointestinal lining. The possibility of restoring function to damaged tissue following disease, injury or ageing depends on its capacity for tissue repair.
TGF-β signalling is associated with the bone morphogenetic protein (BMP) signalling pathway ( Fig. 1.12C ). BMPs are members of the TGF-β superfamily and regulate cell growth, differentiation and development in a wide range of biological processes by activating SMAD transcription factor proteins.
BMP/TGF-β ligands induce the dimerization and then heteromerization of serine/threonine receptor kinases and phosphorylation of the cytoplasmic signalling molecules SMAD2 and SMAD3 for the TGF-β pathway, or SMAD1/5/8 for the BMP pathway. The common SMAD4 transducer translocates to the nucleus. Activated SMADs regulate several biological processes by cell-specific modulation of transcription. TGF-β is a tumour suppressor of premalignant cells but enhances invasion and metastasis of more advanced carcinomas. Mutations of SMAD4 genes are frequent in gastrointestinal and pancreatic tumours. For further reading on cell signalling pathways, see .
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here