Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Pharmacogenetic variants are DNA variants that influence the metabolism and elimination or the action of medications. Research in pharmacogenetics seeks to understand how these variants influence variability in medication response. For the perinatal pharmacologist, genetic variations that affect drug disposition in the fetus, the mother, and the neonate must be considered as one of many sets of variables, along with developmental changes in fetal and neonatal gene expression, organ maturation, placental transfer and metabolism of drugs, and a host of hormonal and environmental influences, which can combine to produce unique patterns of toxicity in mother and child upon medication exposure. Therefore knowledge of potential genetic contributions to drug-induced toxicity in pregnancy, in utero, and in postnatal life can be important in optimizing dosing of drugs, as well as in understanding adverse drug reactions (ADRs) and preventing their further occurrence in patients.
The field of pharmacogenomics is rapidly expanding, and the number of medications with genetic information in the drug label is increasing. Several actionable gene-drug pairs have been clinically implemented in hospitals across the world. Most pharmacogenetic research has been performed in adults, but many of the associations also could apply to children and neonates that are prescribed the same medications.
There are several resources that provide guidance on pharmacogenetic research and implementation. The first step in pharmacogenetics is defining the variants that influence the medication absorption, distribution, metabolism, elimination, or response. The Pharmacogene Variation Consortium ( PharmVar.org ) is a repository of pharmacogene variation that focuses on haplotype structure and allelic variation. Pharmacogenetic alleles can contain one or more variants, and sometimes individual variants are included in the definition of more than one allele. Thus, it is very important to have a common terminology for allele nomenclature, which PharmVar provides. The Pharmacogenomics Knowledge Base ( PharmGKB.org ) is an NIH-funded resource that curates pharmacogenetic literature, including in vitro and in vivo knowledge from medication labels, clinical trials, clinical research, and basic research. Every gene-drug pair in the literature is given a level of evidence based on the association of individual variants with clinical and/or in vitro data. The Clinical Pharmacogenetics Implementation Consortium ( CPIC, cpicpgx.org ) is a group that publishes evidence-based clinical practice guidelines, facilitating the implementation of pharmacogenetics into clinical care. Currently, there are 23 guidelines that impact 49 medications and involve 18 genes ( Table 18.1 ). CPIC utilizes standard nomenclature for allele functionality and activity and provides guidance on electronic health record integration of pharmacogenetic results.
| Drug | Gene(s) | Guideline | References |
|---|---|---|---|
| Abacavir | HLA-B | Do not use in patients with the HLA-B∗57:01 allele | |
| Allopurinol | HLA-B | Do not use in patients with the HLA-B∗58:01 allele | , |
| Amitriptyline | CYP2D6, CYP2C19 | Do not use in CYP2D6 PMs or CYP2D6/CYP2C19 UMs; use 50% lower dose in CYP2C19 PMs | , |
| Atazanavir | UGT1A1 | Do not use in PMs | |
| Atomoxetine | CYP2D6 | Use reduced doses in IMs and PMs | |
| Azathioprine | TPMT, NUDT15 | Extreme dose reduction in TPMT or NUDT15 PMs; reduce dose in TPMT or NUDT15 IMs. | , , |
| Capecitabine | DPYD | Do not use in homozygous DPYD-deficient patients; use 50% of target dose in heterozygotes | |
| Carbamazepine | HLA-B, HLA-A | Do not use in carbamazepine-naïve patients with at least one HLA-B∗15:02 or HLA-A∗31:01 allele | , |
| Citalopram | CYP2C19 | Do not use in CYP2C19 UMs; reduce starting dose by 50% in PMs | |
| Clomipramine | CYP2D6, CYP2C19 | Do not use in CYP2D6 PMs or CYP2D6/CYP2C19 UMs; use 50% lower dose in CYP2C19 PMs | , |
| Clopidogrel | CYP2C19 | Use alternative antiplatelet drugs in CYP2C19 PMs or heterozygotes | , |
| Codeine | CYP2D6 | Do not use in CYP2D6 UMs or PMs | , |
| Desflurane | RYR1, CACNA1S | Do not use in patients with risk variants for malignant hyperthermia | |
| Desipramine | CYP2D6 | Do not use in CYP2D6 UMs or PMs; consider a 25% dose reduction in heterozygotes | , |
| Doxepin | CYP2D6, CYP2C19 | Do not use in CYP2D6 PMs or CYP2D6/CYP2C19 UMs; use 50% lower dose in CYP2C19 PMs | , |
| Efavirenz | CYP2B6 | Decrease dose in CYP2B6 IMs and PMs | |
| Enflurane | RYR1, CACNA1S | Do not use in patients with risk variants for malignant hyperthermia | |
| Escitalopram | CYP2C19 | Do not use in CYP2C19 UMs; reduce starting dose by 50% in PMs | |
| 5-Fluorouracil | DPYD | Do not use in homozygous DPYD-deficient patients; use 50% of target dose in heterozygotes | |
| Fluvoxamine | CYP2D6 | Reduce starting dose by 25%–50% in CYP2D6 PMs | |
| Halothane | RYR1, CACNA1S | Do not use in patients with risk variants for malignant hyperthermia | |
| Imipramine | CYP2D6, CYP2C19 | Do not use in CYP2D6 PMs or CYP2D6/CYP2C19 UMs; use 50% lower dose in CYP2C19 PMs | , |
| Isoflurane | RYR1, CACNA1S | Do not use in patients with risk variants for malignant hyperthermia | |
| Ivacaftor | CFTR | Use only in patients with at least one indicated CFTR variant | |
| 6-Mercaptopurine | TPMT, NUDT15 | Extreme dose reduction in TPMT or NUDT15 PMs; reduce dose in TPMT or NUDT15 IMs. | , , |
| Methoxyflurane | RYR1, CACNA1S | Do not use in patients with risk variants for malignant hyperthermia | |
| Nortriptyline | CYP2D6 | Do not use in CYP2D6 UMs or PMs; consider a 25% dose reduction in heterozygotes | , |
| Ondansetron | CYP2D6 | Do not use in CYP2D6 UMs | |
| Oxcarbazepine | HLA-B, HLA-A | Do not use in carbamazepine-naïve patients with at least one HLA-B∗15:02 or HLA-A∗31:01 allele | , |
| Oxycodone | CYP2D6 | Consider alternative opioids in CYP2C6 UMs and PMs | , |
| Paroxetine | CYP2D6 | Consider other drugs for CYP2D6 UMs and PMs; if paroxetine is warranted, reduce starting dose by 50% | |
| PEG-interferon α-2a | IFNL3 | Consider alternate treatments in unfavorable response genotypes rs12979860 T allele carriers | |
| PEG-interferon α-2b | IFNL3 | Consider alternate treatments in unfavorable response genotypes rs12979860 T allele carriers | |
| Phenytoin | CYP2C9, HLA-B | Reduce dose in CYP2C9 PMs; do not use in patients with the HLA-B∗15:02 allele | |
| Rasburicase | G6PD | Do not use in G6PD-deficient patients | |
| Ribavirin | IFNL3 | Consider alternate treatments in unfavorable response genotypes rs12979860 T allele carriers | |
| Sertraline | CYP2C19 | Reduce starting dose by 50% in CYP2C19 PMs or consider an alternative drug | |
| Sevoflurane | RYR1, CACNA1S | Do not use in patients with risk variants for malignant hyperthermia | |
| Simvastatin | SLCO1B1 | Use lower daily doses or consider alternative therapies in patients with 1 or 2 copies of the rs4149056 C allele | , |
| Succinylcholine | RYR1, CACNA1S | Do not use in patients with risk variants for malignant hyperthermia | |
| Tacrolimus | CYP3A5 | Increase starting dose by 50%–100% in patients who are CYP3A5 EMs or heterozygotes | |
| Tamoxifen | CYP2D6 | Consider alternative therapy in CYP2D6 IMs and PMs | |
| Tegafur | DPYD | Do not use in homozygous DPYD-deficient patients; use 50% of target dose in heterozygotes | |
| Thioguanine | TPMT, NUDT15 | Extreme dose reduction in TPMT or NUDT15 PMs; reduce dose in TPMT or NUDT15 IMs | , , |
| Tramadol | CYP2D6 | Consider alternative opioids in CYP2C6 UMs and PMs | , |
| Trimipramine | CYP2D6, CYP2C19 | Do not use in CYP2D6 PMs or CYP2D6/CYP2C19 UMs; use 50% lower dose in CYP2C19 PMs | , |
| Tropisetron | CYP2D6 | Do not use in CYP2D6 UMs | |
| Voriconazole | CYP2C19 | Do not use in CYP2C19 UMs, RMs, and PMs | |
| Warfarin | CYP2C9, VKORC1, CYP4F2 | Calculate dose based on validated pharmacogenetic algorithms for pediatric patients | , |
Most individuals have at least one pharmacogenetic variant but may not receive the medication whose dosing would be informed by that variant. , This chapter provides an overview and examples of some selected pharmacogenes that influence the pharmacokinetics, pharmacodynamics, and severe adverse reactions to drugs; where possible, these examples were chosen to provide particular relevance to medication use in children.
The most well-established pharmacogenes are those encoding drug-metabolizing enzymes; variants in these genes affect the exposure by altering the rate of metabolism of the drugs. For example, slow metabolism of a pro-drug would result in decreased exposure to the active metabolite and the potential for inefficacy, but slow metabolism of an active drug into an inactive compound would result in increased exposure to the active drug and the potential for toxicity. The opposite is true for faster metabolism due to genetic variants. For pharmacogenes that influence pharmacokinetics, often the recommendations are to adjust dosage, but for some medications, an alternative medication is recommended. There are two phases of drug metabolism—the first is oxidation or hydrolysis, and the second is conjugation (e.g., glucuronidation). Many drugs are metabolized by oxidation through the cytochrome P450 family of enzymes expressed highly in the liver ( Fig. 18.1 ).
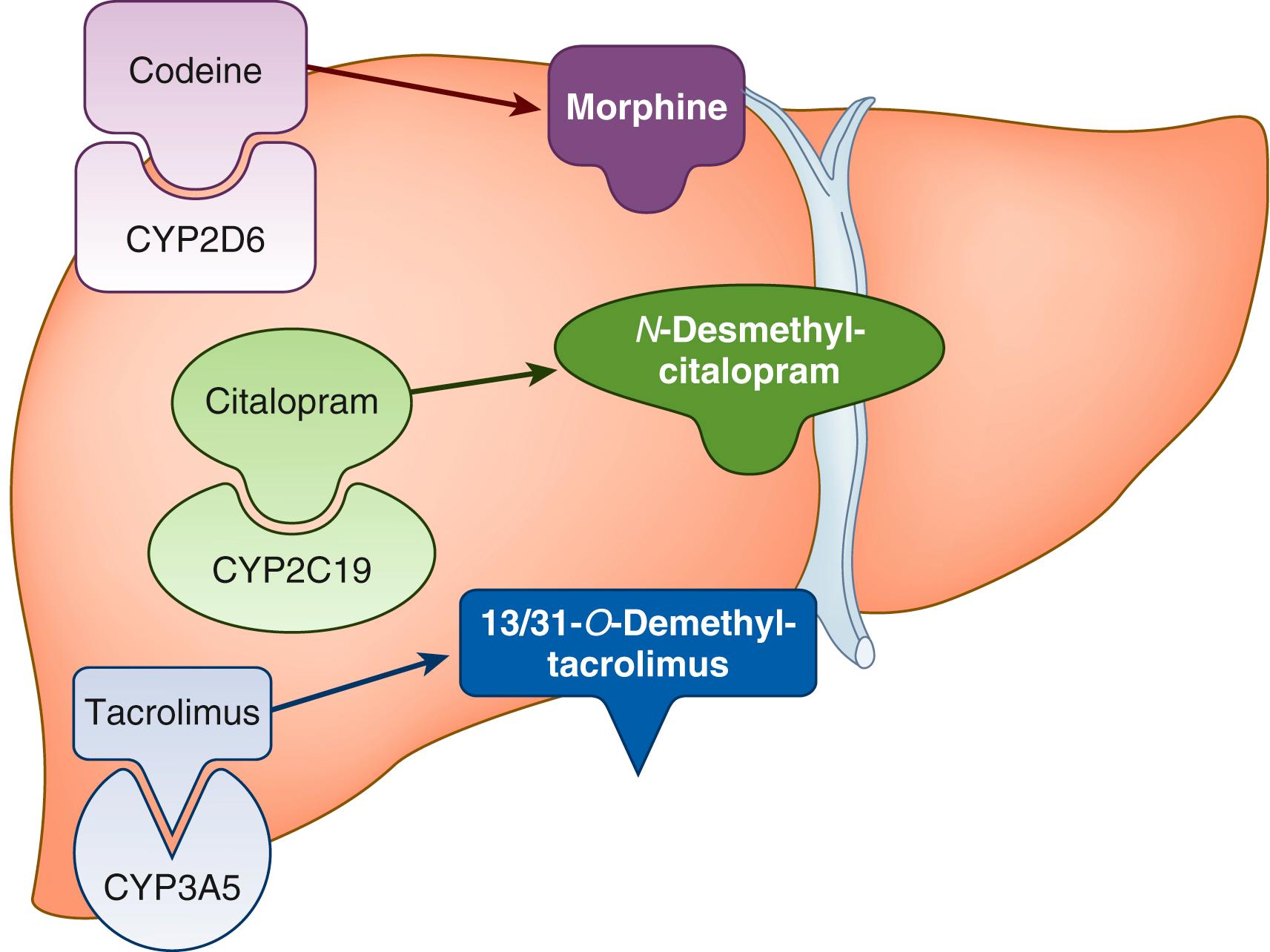
The first pharmacogene characterized was CYP2D6 , and to date, more than 130 alleles have been described for this gene ( https://www.pharmvar.org/gene/CYP2D6 ). Approximately 25% of all medications are metabolized by the CYP2D6 enzyme to some degree, and this gene is included on several drug labels and 6 CPIC guidelines, with dosing adjustments or changes in medication recommended. Testing of CYP2D6 is required by the US Food and Drug Administration (FDA) prior to initiation of three medications: eliglustat, pimozide, and tetrabenazine ( Table 18.2 ).
| Drug | Therapeutic Area | Biomarker |
|---|---|---|
| Isosorbide Dinitrate | Cardiology | CYB5R |
| Isosorbide Mononitrate | Cardiology | CYB5R |
| Nebivolol | Cardiology | CYP2D6 |
| Propafenone | Cardiology | CYP2D6 |
| Metoclopramide (2) | Gastroenterology | G6PD |
| Metoclopramide (3) | Gastroenterology | CYP2D6 |
| Warfarin (1) | Hematology | CYP2C9 |
| Warfarin (2) | Hematology | VKORC1 |
| Carglumic Acid | Inborn errors of metabolism | NAGS |
| Eliglustat | Inborn errors of metabolism | CYP2D6 |
| Migalastat | Inborn errors of metabolism | GLA |
| Sodium Phenylbutyrate | Inborn errors of metabolism | ASS1, CPS1, OTC (urea cycle disorders) |
| Abacavir | Infectious diseases | HLA-B |
| Dapsone (3) | Infectious diseases | G6PD |
| Primaquine (1) | Infectious diseases | G6PD |
| Tafenoquine | Infectious diseases | G6PD |
| Amifampridine | Neurology | NAT2 |
| Amifampridine Phosphate | Neurology | NAT2 |
| Clobazam | Neurology | CYP2C19 |
| Deutetrabenazine | Neurology | CYP2D6 |
| Siponimod | Neurology | CYP2C9 |
| Tetrabenazine | Neurology | CYP2D6 |
| Valbenazine | Neurology | CYP2D6 |
| Ado-Trastuzumab Emtansine | Oncology | ERBB2 (HER2) |
| Afatinib | Oncology | EGFR |
| Alectinib | Oncology | ALK |
| Alpelisib (1) | Oncology | ERBB2 (HER2) |
| Alpelisib (2) | Oncology | ESR (hormone receptor) |
| Alpelisib (3) | Oncology | PIK3CA |
| Atezolizumab (1) | Oncology | CD274 (PD-L1) |
| Belinostat | Oncology | UGT1A1 |
| Binimetinib (1) | Oncology | BRAF |
| Bosutinib | Oncology | BCR-ABL1 (Philadelphia chromosome) |
| Brentuximab Vedotin (2) | Oncology | TNFRSF8 (CD30) |
| Ceritinib | Oncology | ALK |
| Cetuximab (1) | Oncology | EGFR |
| Cetuximab (2) | Oncology | RAS |
| Cobimetinib | Oncology | BRAF |
| Crizotinib (1) | Oncology | ALK |
| Crizotinib (2) | Oncology | ROS1 |
| Dabrafenib (1) | Oncology | BRAF |
| Dabrafenib (3) | Oncology | RAS |
| Dacomitinib | Oncology | EGFR |
| Dasatinib | Oncology | BCR-ABL1 (Philadelphia chromosome) |
| Enasidenib | Oncology | IDH2 |
| Encorafenib | Oncology | BRAF |
| Erdafitinib (1) | Oncology | FGFR |
| Erlotinib | Oncology | EGFR |
| Everolimus (1) | Oncology | ERBB2 (HER2) |
| Everolimus (2) | Oncology | ESR (hormone receptor) |
| Exemestane | Oncology | ESR, PGR (hormone receptor) |
| Gefitinib (1) | Oncology | EGFR |
| Gilteritinib | Oncology | FLT3 |
| Imatinib (1) | Oncology | KIT |
| Imatinib (2) | Oncology | BCR-ABL1 (Philadelphia chromosome) |
| Imatinib (3) | Oncology | PDGFRB |
| Imatinib (4) | Oncology | FIP1L1-PDGFRA |
| Irinotecan | Oncology | UGT1A1 |
| Ivosidenib | Oncology | IDH1 |
| Lapatinib (1) | Oncology | ERBB2 (HER2) |
| Lapatinib (2) | Oncology | ESR, PGR (hormone receptor) |
| Larotrectinib | Oncology | NTRK |
| Mercaptopurine (1) | Oncology | TPMT |
| Mercaptopurine (2) | Oncology | NUDT15 |
| Midostaurin (1) | Oncology | FLT3 |
| Nilotinib (1) | Oncology | BCR-ABL1 (Philadelphia chromosome) |
| Olaparib (1) | Oncology | BRCA |
| Olaparib (2) | Oncology | ERBB2 (HER2) |
| Osimertinib | Oncology | EGFR |
| Panitumumab (2) | Oncology | RAS |
| Pembrolizumab (2) | Oncology | CD274 (PD-L1) |
| Pembrolizumab (3) | Oncology | Microsatellite instability, mismatch repair |
| Pertuzumab (1) | Oncology | ERBB2 (HER2) |
| Rituximab | Oncology | MS4A1 (CD20 antigen) |
| Rucaparib (1) | Oncology | BRCA |
| Talazoparib (1) | Oncology | BRCA |
| Thioguanine (1) | Oncology | TPMT |
| Thioguanine (2) | Oncology | NUDT15 |
| Trametinib (1) | Oncology | BRAF |
| Trastuzumab (1) | Oncology | ERBB2 (HER2) |
| Vemurafenib (1) | Oncology | BRAF |
| Aripiprazole | Psychiatry | CYP2D6 |
| Aripiprazole Lauroxil | Psychiatry | CYP2D6 |
| Atomoxetine | Psychiatry | CYP2D6 |
| Brexpiprazole | Psychiatry | CYP2D6 |
| Citalopram (1) | Psychiatry | CYP2C19 |
| Clozapine | Psychiatry | CYP2D6 |
| Iloperidone | Psychiatry | CYP2D6 |
| Pimozide | Psychiatry | CYP2D6 |
| Vortioxetine | Psychiatry | CYP2D6 |
| Azathioprine (1) | Rheumatology | TPMT |
| Azathioprine (2) | Rheumatology | NUDT15 |
| Celecoxib | Rheumatology | CYP2C9 |
One example of a medication metabolized by this gene with clinical importance to pediatric patients is codeine, which is metabolized to the active drug (morphine) by CYP2D6. Individuals with little or no CYP2D6 activity (poor metabolizers) receive inadequate exposure to morphine with conventional dosing and need alternative analgesic medications to control pain (see Table 18.2 ). , Alternatively, those with duplication of the CYP2D6 gene (ultrarapid metabolizers) are at risk for sedation and respiratory depression from generating high concentrations of morphine. Since the FDA issued a contraindication for codeine use in children younger than 12, many hospitals removed codeine from their formularies or restricted its use. The FDA also issued a warning to mothers that breast-feeding is not recommended when taking codeine due to the risk of serious adverse reactions (excess sleepiness, difficulty breast-feeding, or breathing problems). However, the American College of Obstetricians and Gynecologists does not recommend against using codeine in breast-feeding mothers but recommends counseling mothers on risks and newborn signs of toxicity if codeine-containing medications are selected for postpartum pain control.
Another pharmacogene that encodes an enzyme that metabolizes many commonly prescribed medications is CYP2C19 . This gene has alleles with increased function (∗17), decreased function, and no function; therefore phenotypes can be ultrarapid, rapid, normal, intermediate, or poor metabolizer. The medications metabolized by CYP2C19 include antidepressants (e.g., escitalopram, sertraline, amitriptyline), clopidogrel, voriconazole, and proton pump inhibitors. Testing of this gene is not required prior to initiating any medications, but it is included in the FDA labels of 22 medications ( www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling ), usually in the Clinical Pharmacology section, but for citalopram and clobazam, it is included in the dosage and administration section, where dose reductions are recommended in poor metabolizers to avoid adverse reactions (see Table 18.2 ).
Tacrolimus is an immunosuppressant that is metabolized by CYP3A5. Variants in CYP3A5 (e.g., the ∗3 allele) explain 40% to 50% of the variability in blood concentrations of tacrolimus. , This is one of the few gene-drug pairs that has been tested in randomized clinical trials to test conventional dosing versus genotype-guided dosing. A trial in pediatric patients receiving a solid organ transplant demonstrated the time to the therapeutic concentration was reached sooner in a genotype-guided group than an unguided group and there were no differences in adverse events. Since CYP3A5 is expressed highly in the liver, in patients receiving a liver transplant, the donor liver must be genotyped in order to provide genotype-guided dosing in these patients.
Thiopurines are metabolized by thiopurine methyltransferase (TPMT) into inactive metabolites. Patients receiving normal doses of thiopurines that are TPMT poor metabolizers are at very high risk for acute toxicity and require markedly reduced doses (10-fold lower). TPMT variants account for much of the variability in thiopurine intolerance in people of European and African ancestry; however, variants in another gene in the thiopurine metabolism pathway, NUDT15 , accounts for the majority of the variability in thiopurine intolerance in people of Asian ancestry and have also been found in Hispanic patients. Patients with NUDT15 no-function alleles also require drastic dose reductions in thiopurines to avoid severe myelosuppression. Adjusting dosages of thiopurines based on TPMT genotype has reduced the incidence of adverse effects without compromising efficacy.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here