Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
This work was supported in part by an award from Soy Health Research Program (SHRP, United Soybean Board, Chesterfield, MO, USA), University of South Carolina School of Medicine Research Development Fund (USC SOM RDF, Columbia, SC, USA), South Carolina Spinal Cord Injury Research Fund (SC SCIRF-2015-I-01, Columbia, SC, USA), and earlier grants (R01 CA91460 and R01 NS057811) from the National Institutes of Health (Bethesda, MD, USA).
The small non-coding RNAs with an average length of 22 nucleotides are known as microRNAs or miRNAs, which are evolutionarily conserved and found only in eukaryotes including humans. The first miRNA was discovered 25 years ago in Caenorhabditis elegans , showing the complementarity of this miRNA to the 3′ untranslated region (3′UTR) of a target messenger RNA (mRNA) for RNA-RNA interaction for degradation (silencing) of the target mRNA . Since then we have learned a lot about the biogenesis of miRNAs, their structures, characteristics, and roles in regulating expression of genes in normal cells as well as in cancer cells. Biogenesis of more than 1000 novel miRNAs has been discovered in humans . It has been predicted that miRNAs account for about 5% of the human genome and control at least 30% of protein-coding genes. Endogenous individual genes encode intergenic miRNAs while introns of protein-coding genes encode intronic miRNAs . Most of the miRNA genes (60%) are intergenic or oriented antisense to the neighboring genes and thus can be transcribed independently. About 40% of miRNA genes are found in introns of protein-coding and nonprotein-coding genes. It has also been reported that occasionally a miRNA gene may be transcribed in concert with its host protein-coding gene, making the process of transcription a concurrent regulation of the miRNA and protein-coding gene.
All miRNAs are single-stranded RNA molecules (ssRNA) with a stem-loop structure, highly stable, partially complementary to the 3′UTR of the targets, and able to act on multiple targets. Since most sites on the target mRNA show only partial complementarity with their matching miRNA, individual miRNAs can destroy as many as 100 different target mRNAs. Further, a specific mRNAs may contain multiple binding sites for various miRNAs, producing a network of complex regulation. Because miRNAs are highly conserved molecules, they must have highly regulatory roles in activation or inhibition of many normal cell signaling pathways, deregulation of which can cause diseases including cancers. Recent research highlights the refined molecular mechanisms of miRNAs in the transcriptional regulation of the target genes and their high potential as biomarkers and therapeutics in human malignant neuroblastoma.
Currently, specific miRNAs are used as biomarkers in human cancers . A wide range of miRNAs is known to be involved in progression or suppression of tumor growth. So, it is important to know how miRNAs are named for their very specific identification . As a standard process, a name is given to the experimentally confirmed new miRNA before its publication. The prefix “miR” refers to pri-miRNA and the pre-miRNA. The name of a mature miRNA includes the prefix “miR” followed by a hyphen and a number (e.g., miR-123), where the number indicates the order of naming. Two miRNAs with almost identical sequences except in one or two nucleotides are marked with an additional lowercase letter (e.g., miR-123a, miR-123b). Two mature miRNAs derived from opposite arms of the same pre-miRNA are designated with a suffix -3p or -5p (e.g., miR-123-3p, miR-123-5p). If relative levels of expression of two miRNAs are known, an asterisk after the name indicates the miRNA that is expressed at a low level relative to the other miRNA in the opposite arm of a hairpin (e.g., miR-123∗ is low expressing relative to miR-123). Further, a recent report suggests that there is a need for a revision of miRNA nomenclature and terminology to include freshly identified miRNA variants, organellar miRNAs, novelty of miRNAs, evolutionary history or biogenesis of miRNAs, and the confidence in their identification .
Aberrant expression of miRNAs has been implicated in tumorigenesis. Currently, clinical diagnosis of human malignant neuroblastoma is based on various genetic anomalies and prospective epigenetic variations . Also, the determination of aberrant expression of miRNAs is highly useful in branding the pathogenesis and heterogeneity of a specific neuroblastoma . Because we still lack a complete understanding of the intricacy of miRNA networks that cause pathogenesis in neuroblastoma, we continue to face multiple difficulties in identifying novel miRNA biomarkers or therapeutic targets for successful treatment of malignant neuroblastoma . Recent findings show that several oncogenic miRNAs are upregulated and some tumor suppressive miRNAs are downregulated for pathogenesis in human malignant neuroblastoma . Per se research on miRNAs will continue to provide us with useful biomarkers for diagnosis, risk stratification, treatment, and prognosis of human malignant neuroblastoma. Inhibition of expression of specific oncogenic miRNAs, which cause tumor progression and drug resistance, and induction of expression of specific tumor suppressor miRNAs, which promote cell cycle arrest and apoptosis, may give us potential therapeutic strategies for controlling the growth of human malignant neuroblastoma.
Many different regions of the human genome harbor miRNA genes, which are commonly found in clusters. The miRNA genes most frequently occur in intergenic regions and introns of protein-coding genes. Previously these regions were considered as “junk DNA” in the human genome due to their unknown biological functions. However, we now know that these are not “junk DNA” after all. These are instead jewel DNA regions that host the majority of the miRNA genes in our genome. miRNA genes are rarely found in exons of protein-coding genes and antisense DNA strands . Transcription and regulation of miRNA genes vary depending on their loci. Transcription of an intronic miRNA gene located on a host gene with the same orientation occurs in consort with the host gene by the same promoter . On the contrary, intergenic miRNA genes have their own promoters and produce long polycistronic transcripts that resemble mRNA as they contain distinct 5′ and 3′ boundaries and poly(A) tails . Majority of miRNA genes are transcribed by RNA polymerase II (pol II) . However, some miRNA genes are transcribed by RNA pol III that specifically produces small nonprotein-coding RNAs for regulating cell cycle and growth .
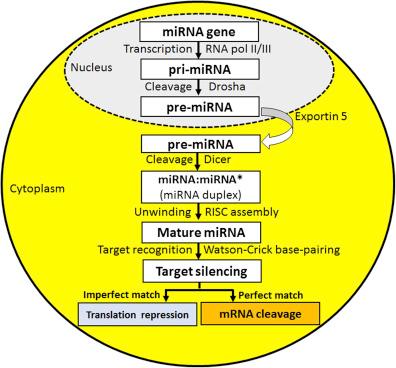
Biogenesis and maturation of human miRNAs occur in four steps: transcription, nuclear processing, nuclear exportation, and cytoplasmic processing . The miRNA gene is transcribed to a primary miRNA (pri-miRNA), which is processed into precursor miRNA (pre-miRNA) and then miRNA duplex (miRNA:miRNA∗, where the asterisk indicates passenger strand) for the final release of mature miRNA ( Fig. 17.1 ). Activity of RNA pol II/III on the miRNA gene produces a several kb long primary miRNA (pri-miRNA), which assumes a stem-loop structure for recognition and cleavage by Drosha (a nuclear RNA III endonuclease) to generate a 60–70 nucleotides long precursor miRNA (pre-miRNA) for exportation via Exportin 5 to the cytoplasm, where the pre-miRNA is further processed by Dicer (another RNA III endonuclease) to the final ∼22 nucleotides long miRNA duplex for attachment of 5′ end of its strand through the weakest base pairing to the RNA-induced silencing complex (RISC) and movement to the 3′UTR of the target mRNA . About 45% of all miRNAs are originated from the miRNA genes containing a common promoter and polycistronic units with multiple discrete loops from which mature miRNAs are produced. A single miRNA is capable of suppressing the expression of multiple mRNAs directly or indirectly, suggesting that inhibition or induction of expression and function of even a single miRNA in cancers can have profound consequences in cell signaling pathways. The principal mechanism of action of a miRNA is the inhibition of expression of the target gene or genes; however, in rare cases, miRNAs may modulate transcription or activate translation . Because miRNAs in almost all cases kill the mRNAs, they are readily recognized as the negative regulators of the gene expression.
Recent results reported from various laboratories suggest that miRNAs are the master regulators of intracellular signaling pathways in cancers . They have specific targets and play important roles in cell cycle, proliferation, differentiation, apoptosis, and metabolism in normal and cancer cells . Hence, it is not surprising that aberrant expression of miRNAs is associated with the development of many cancers, production and survival of cancer stem cells, hypoxia, induction of autophagy, development of multidrug resistance, occurrence of epithelial-mesenchymal transition (EMT), cell migration, cell invasion, promotion of angiogenesis, and tumor metastasis . A prerequisite for tumor metastasis is EMT, which is a process for making the epithelial cells lose their cell-to-cell contact proteins such as E-cadherin and γ-catenin for conversion to mesenchymal cells that express vimentin and fibronectin . After the EMT event, tumor cells gain several advantages that include their newly found ability to separate from the primary tumor and thus gain complete freedom of movement, leading to tumor metastasis. As mentioned earlier, depending on their targets, a miRNA can act as an oncogene and/or tumor suppressor to control tumorigenesis. The principal function of the oncogenic miRNAs is the elimination of the mRNAs of tumor suppressor genes, while that of the tumor suppressor miRNAs is the destruction of the mRNAs of oncogenes.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here