Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The authors wish to thank Jenny L. Heidbrink, Victoria Bushman, Erin Brand, Jim Norton, Jun Kim, Brian Feild, Roxanne Armstrong, Henrik Olsen, Will Fitzhugh, Jim Duff, Ping Zhang and Katherine Paweletz, for their technical assistance. We also wish to thank Samuel Broder, Robert Booth and Scott Patterson for their advise on this project.
Unlike most other cancers, incidences of pancreatic cancer have been on the rise . It is the fourth leading cause of cancer death in the United States; in 2013, an estimated 45,220 men and women will be diagnosed with pancreatic cancer, and 38,460 will die from this disease . Pancreatic cancer is a challenging disease as symptoms experienced by patients are often nonspecific. Since there are no early detection tools available, patients are often diagnosed at a late stage. As such, even with today’s best treatment efforts, only 5% of pancreatic cancer patients will survive 5 years beyond their initial diagnosis .
With only 15–20% of cases deemed to be operable, the standard of care for nonresectable patients remains to be treatment with the chemotherapeutic gemcitabine . It is discouraging to note that gemcitabine provides a median survival rate of only 6 months. Though a number of alternative treatment options have been in clinical testing, the majority of these have failed to demonstrate improvements. The most promising so far is FOLFIRINOX, a chemotherapeutic combination (oxaliplatin, irinotecan, fluorouracil, and leucovorin) that has been proven under phase III clinical trial setting to extend overall survival rate to 11.1 months . Whilst this result is encouraging, it is clear that there is an urgent need for the identification of new diagnostic and therapeutic strategies for this disease.
In recent years, there has been increasing interest in targeted approaches for cancer. For certain types of cancers, such as breast cancer , chronic myeloid leukemia , lymphomas , and non-small cell lung cancer , this approach has been proven to be very effective. For pancreatic cancer, development of targeted therapy has been more challenging . Genetically, pancreatic ductal adenocarcinoma (PDA, the most common form of pancreatic cancer), universally carry one of four defects: activating KRAS2 oncogene (70–90% of tumors), inactivation of p16/CDKN2A gene (75–80% of tumors), TP53 abnormality (50–75% of tumors), and deletion in SMAD4 gene (50–60% of tumors) . Unfortunately, none of these mutations is targetable by the two approaches that have proven to be successful for cancer treatment: targeted small molecules and antibody-based therapies. Whilst strategies that target druggable enzymes downstream of these modifications are being pursued, they have so far been ineffective in the clinical setting. For example, inhibitors for farnesyltransferase, an enzyme that is important for Ras activation, demonstrated no survival benefit for pancreatic cancer patients when compared to standard therapy .
Cell surface proteins account for more than two-thirds of known drug targets . These proteins are easily accessible and play significant, functional roles in tumorigenesis and are involved in processes such as cell proliferation, cell adhesion, and cell invasion. The identification of cell surface proteins that specifically target pancreatic cancer may therefore yield novel treatment targets for this disease. However, due to their hydrophobic nature and low abundance, it has been difficult to identify new candidates for drug therapy.
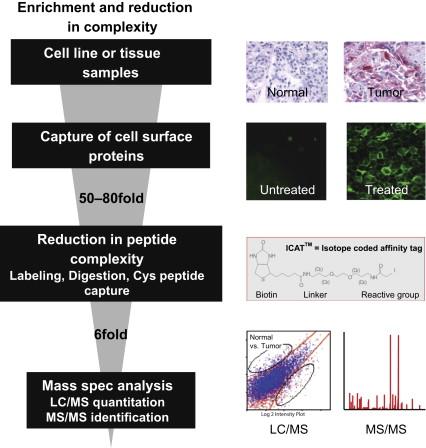
Proteomics is a technique that offers the opportunity to identify hundreds of differentially expressed proteins. This often leaves the researcher with the daunting task of having too many potential targets to validate . One of the goals in our laboratory is to reduce sample complexity so the proteomic can be performed at greater depth. This enabled us to obtain a focused list of differentials, each of which can then be thoroughly validated using an array of well-defined expression and functional validation methods . To this end, we isolated cell surface membrane proteins from cell lines and performed LC-MS/MS on this specific subcellular compartment . The expression validation platform and the RNAi target validation platform that was utilized to assign roles these proteins play in pancreatic tumorigenesis will be described. Example proteins that were identified and validated using this platform, namely Tissue Factor, Integrin Beta 6, and Na + /K + ATPase Beta 3, will be presented. We provide here a first report of identification and validation of Na + /K + ATPase Beta 3 subunit as a pancreatic cancer target. We believe this strategy, as outlined in Figure 11.1 , has allowed us to identify potentially valuable therapeutic and diagnostic targets.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here