Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
This work was supported in part by the National Institutes of Health under grants K25CA137222, R21CA149772, R21CA161575, and R01CA107209, and Canary Foundation.
Despite advances in our understanding of pancreatic cancer development in the past decade, the disease remains the fourth most common cause of cancer death in the United States . Unfortunately, its 5-year survival rate has not been improved significantly in the past 30 years. The major reason for this poor prognosis is that the majority of pancreatic cancer patients are diagnosed at a late stage with metastatic, inoperable disease, in which no effective treatments are currently available. Even for the small group of patients diagnosed with resectable cancer and who do undergo surgical resection, the 5-year survival rate approximates 15–40% . Methods for earlier detection and more effective therapeutic treatments are much needed to improve the clinical outcome of pancreatic cancer.
Proteins are the essential molecules that regulate and participate in biological functions. Proteome alterations that are associated with diseases may include changes in protein expression, post-translational modifications (PTMs) and protein–protein interactions, which may all lead to malfunction of cellular biological processes. Identification and quantification of protein abnormalities associated with pancreatic cancer pathways, thus, may supply molecular information that provides new disease surrogates for diagnosis or novel therapeutic targets. In some cases, identification of a key regulatory protein could provide information for both diagnostics and treatment, known as theranostics.
Over the past decade, researchers have looked beyond the scope of genomics to explore protein-driven functional changes that are associated with pancreatic tumorigenesis. Advances in proteomics, especially quantitative proteomics, enables systematic investigation of malignancy-related proteome alterations that affect cellular physiology and function. This type of information can provide for new hypotheses bridging the gap between basic biological understanding and translational research. As illustrated in Figure 9.1 , clinical specimens for proteomics investigation can include pancreatic tissues, plasma/serum, pancreatic juice, and cyst fluids, as well as isolated cells, from individuals who are in good health and those with cancer or other pancreatic diseases. Such samples are investigated to identify signaling pathways and molecular events underlying pancreatic tumorigenesis—laying a foundation for translational exploration of key proteins to improve patient care.
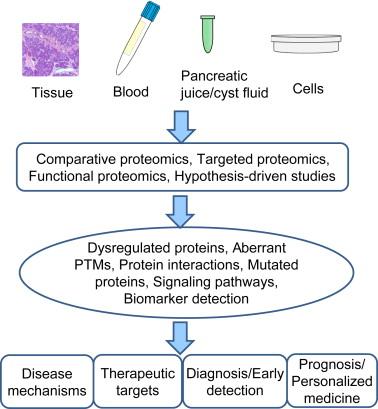
A major challenge in proteomics analysis arises from the enormous complexity of protein constituents and the vast dynamic range in protein abundance in biological samples. Comprehensive interrogation of a protein profile, particularly of low-abundant proteins in a complex biological system that may consist of thousands or more protein species with the addition of their isoforms, PTMs, mutations, and polymorphisms is a dedicated task and requires a concerted approach drawn from different techniques. A typical proteomics pipeline requires four technical modules, including sample preparation, protein/peptide separation, mass spectrometric analysis, and bioinformatics. In addition, shotgun proteomics-based quantitative analysis entails a variety of stable isotope-labeling methodologies that can be used to introduce differential mass tags on proteins or peptides to facilitate the quantitative analysis.
In large-scale proteomics profiling experiments, effective fractionation and separation of proteins and peptides before mass spectrometric analysis enhances the analytical range and capacity. The nature of the biological specimens and experimental design can influence the approach used to prepare proteins for analysis. For gel-based quantitative analysis, two-dimensional electrophoresis (2-DE) is the most common way to separate proteins in a complex biological sample, and the staining intensities of proteins are used to achieve comparative quantification . The identification of selected protein spots is accomplished through mass spectrometric analysis, typically via in-gel digestion of proteins. Alternatively, shotgun proteomics-based approaches rely on analysis and assignment of peptides for protein identification. Because mass spectrometric analysis at the peptide level provides substantially better analytical sensitivity and mass accuracy compared with direct protein analysis, shotgun proteomics permits sophisticated amino acid sequence identification using an automatic database search. Digestion of a vast number of proteins in a biological sample, however, can generate numerous peptide species with significant dynamic differences in abundance, unavoidably multiplying the complexity of the sample for analysis. In such a setting, a variety of separation techniques have been utilized to effectively separate or fractionate a complex biological sample at either the protein or peptide level before the mass spectrometric interrogation. Typically, orthogonal mechanisms are coupled to maximize the separation efficiency. Proteins can be separated using electrophoresis, liquid chromatography (LC), or size exclusion before enzymatic digestion, whereas at the peptide level, 2D LC is commonly used for peptide fractionation with the combination of ion chromatography and reverse-phase LC, such as is used in the multi-dimensional protein identification technology (MudPIT) .
Differential stable isotopic labeling is the most common and versatile approach for quantitative proteomics analysis. This technique provides mass tags, which allow mass spectrometry to distinguish peptides with identical sequence but from different sample origins (e.g., diseased cases versus healthy controls) for quantitative comparison. There are different ways to incorporate stable isotope labeling onto proteins or peptides, including chemical derivatization and metabolic and enzymatic labeling . Chemical derivatization is the most widely used methodology for stable isotope labeling and can be categorized into two types, isotopic and isobaric, based on how the isotopic signals are generated in mass spectrometric analysis. Isotopic-type stable isotope labeling methods, such as isotope-coded affinity tags (ICAT) and isotope-coded protein label (ICPL) , quantify peptides at the MS level, while isobaric types, such as isobaric tags for relative and absolute quantitation (iTRAQ) and tandem mass tags (TMT) , allow the mass spectrometer to generate and acquire differential isotopic reporting peaks at the tandem mass spectrometry (MS/MS) level after collision-induced dissociation (CID). Chemical derivatization approaches are post protein isolation methods that are well suited for analyzing almost any sample types, including clinical specimens such as tumor tissue, plasma/serum, and pancreatic juice. Metabolic incorporation of stable isotope labeling, such as stable isotope labeling by amino acids in cell culture (SILAC) , utilizes cell culturing to introduce isotopic tags on proteins by supplementing stable isotopic-labeled amino acid in cell culture medium and is particularly suitable for cell model studies. The enzymatic method, such as O 18 labeling , takes a different route to introduce isotopic labeling on peptides by enzymatic digestion of proteins in an O 18 -enriched buffer. Last, with advances in high-resolution mass spectrometric instrument and bioinformatics, label-free-based quantitative proteomics, based on spectral count or signal intensity, also have been applied in a variety of studies .
Mass spectrometer is the center of proteomics analysis, and its major components consist of the ion source, mass analyzer, and detection unit. Ion plume is produced and introduced into mass spectrometer through ion source; the ions are then separated in mass analyzer under an ultra-high vacuum based on their mass-to-charge value and recorded by a detector. The most commonly used ion source techniques are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI), both of which are used widely in proteomics analysis . Several types of mass analyzers are available, including ion trap, orbitrap, time-of-flight (TOF), and quadrupole. These mass analyzers can be used independently or can be combined to achieve a tandem analysis. Recently, an ion mobility technique has been introduced, providing an additional dimension for resolving a complex biological sample within a mass spectrometer. In MS/MS analysis, peptides are fragmented in a collision cell and the fragmentation pattern of each of the peptide is used for sequence identification. In addition to the most commonly used CID mechanism, other soft ionization methods, including electron-transfer dissociation (ETD) and electron-capture dissociation (ECD) have been developed to facilitate protein PTM analysis .
The last module of a proteomics pipeline is bioinformatics data processing, which includes a series of integrated software, from data format conversion to peptide/protein sequence assignment, statistical validation, and quantitative analysis. The MS/MS spectra generated are searched against a protein database for peptide/protein sequence identification using database searching algorithms, such as SQUEST , MASCOT , and X!tandem , followed by statistical validation and false discovery assessment. For quantitative experiments, based on peptide/protein registry, analytical software is used to compare the differential signal intensities between the sample and the control to obtain quantitative information reflecting peptide/protein relative abundance in the samples compared.
In addition to nonbias quantitative proteomics methods for global protein profiling, targeted proteomics utilizes the concept of isotope dilution and provides highly specific and sensitive detection of candidate analytes in a complex biological sample. This approach is increasingly applied in translational and biomarker studies . The most widely used mass spectrometry technique for targeted proteomics analysis is the triple quadrupole-based selected (or multiple) reaction monitoring (SRM or MRM) technique. The development of targeted proteomics technology has opened a new avenue for quantitative detection of targeted proteins, peptides, or even specific forms of PTMs in a complex biological system, carrying great promise to facilitate biomarker development for pancreatic cancer and other diseases.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here