Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Bacteria can be categorized based on the Gram stain reaction (gram-positive or gram-negative), shape (cocci, bacilli, coccobacilli, spirochete), preferred atmosphere (aerobic, microaerophilic, anaerobic), and presence or absence of spores. They can be identified on the basis of key biochemical tests, antigenic components (e.g., cell wall antigens, toxins), and/or molecular features, including nucleic acid–based testing and proteomic detection.
Among the gram-positive cocci, the most important human pathogens (and the infections that they commonly cause) are Staphylococcus aureus (skin and soft-tissue infections, bacteremia, septic arthritis), Streptococcus pyogenes (pharyngitis and its nonsuppurative complications, skin and soft-tissue infections), Streptococcus agalactiae (neonatal bacteremia and meningitis), Streptococcus pneumoniae (community-acquired pneumonia, meningitis), and Enterococcus faecalis and E. faecium (nosocomial urinary tract infections and bacteremia).
Among gram-positive bacilli, the most important human pathogens (and the infections that they commonly cause) are Listeria monocytogenes (meningitis, bacteremia), Nocardia species (pneumonia, soft-tissue infections, brain abscess), Bacillus anthracis (skin and soft-tissue infections, pneumonia; a bioterrorism agent), and Corynebacterium diphtheriae (diphtheria), which is rarely encountered in the clinical laboratory in the United States.
Among gram-negative cocci, the most important human pathogens (and the infections that they commonly cause) are Neisseria meningitidis (meningitis), Neisseria gonorrhoeae (gonorrhea), and Moraxella catarrhalis (upper and lower respiratory tract infections).
Gram-negative bacilli include Enterobacterales, many of which are normal flora in the gastrointestinal tract; nonfermentative gram-negative bacilli (e.g., Pseudomonas aeruginosa and Acinetobacter baumannii ), which are found in the environment and cause human infection when host defenses are compromised; halophilic organisms ( Vibrio spp.); microaerophilic bacteria ( Campylobacter , Helicobacter ); fastidious organisms ( Legionella spp., Bordetella spp., Francisella tularensis , Brucella spp., Haemophilus species); and miscellaneous infrequently encountered bacteria.
Among the Enterobacterales, the most important human pathogens (and the infections that they commonly cause) are Escherichia coli (urinary tract infection, diarrhea, bacteremia), Klebsiella pneumoniae and Klebsiella oxytoca (urinary tract infection, pneumonia, bacteremia), Proteus spp. (urinary tract infection, wound infections), Salmonella spp. (diarrhea, typhoid fever), Shigella spp. (diarrhea), and Enterobacter spp. (nosocomial pneumonia, urinary tract infection, bacteremia).
Among the anaerobes, the most important human pathogens (and the infections that they commonly cause) are the Bacteroides fragilis group (intraabdominal infections, abscesses); Clostridium spp., especially Clostridium perfringens (soft-tissue infections, food poisoning), Clostridium tetani (tetanus), and Clostridioides difficile (antibiotic-associated diarrhea); and non–spore-forming gram-positive anaerobes, such as Actinomyces israelii and Cutibacterium acnes.
A wide variety of bacterial species may be recovered from clinical specimens. To appropriately assess the clinical significance of these organisms, an understanding of the normal bacterial flora present at different anatomic locations is essential. In some cases, the number of organisms present can be extremely high—for example, 10 6 organisms/cm 2 of skin, 10 9 organisms/mL of oral secretions, and 10 11 organisms/g of colon contents. It is important to obtain samples with minimal contamination from the normal flora (Miller et al., 2007; ). This may be difficult but can be optimally achieved if proper procedures are followed. These procedures, along with processing techniques that serve to enhance recovery of pathogenic microorganisms, are discussed in Chapter 66 .
Direct examination of a specimen with a Gram stain is one of the most valuable procedures performed by the microbiology laboratory. The Gram stain result rapidly provides information that is used by the clinician for selecting appropriate antimicrobial therapy. It also helps the clinical laboratory scientist assess the quality of the specimen and the extent to which certain organisms recovered in culture will be worked up. Organisms present in abundant quantity in specimens containing many white blood cells are given more attention than those that are present in smaller numbers in the absence of white blood cells. Multiple specimens positive for similar organisms in smears and cultures contribute to increased clinical significance of the results.
To prepare a smear for staining, an aliquot of the most purulent or bloody portion of the specimen is placed on a clean microscopic slide in a manner that provides both thick and thin areas. For sterile body fluids such as cerebrospinal fluid (CSF), a cytocentrifuge should be used to concentrate the specimen by 10 to 100 times ( ). The material on the slide is allowed to air-dry, is fixed with methanol or gentle heat, and then is stained with Gram stain reagents (crystal violet, Gram iodine, alcohol, and safranin). Organisms that have a gram-positive cell wall will resist decolorization with methanol and will retain the purple color of the crystal violet. Organisms that have a gram-negative cell wall will be decolorized and will stain red with safranin counterstain.
Stained smears are initially examined using a low-power objective to look for large structures, such as nematode larvae, Curschmann spirals, large granules, grains, bacterial microcolonies, or fungal forms. An oil immersion lens is then used to assess the type of bacteria present. Because 10 5 organisms/mL must be present to see one organism per oil immersion field (1000×), smears must be examined carefully to detect small numbers of organisms.
The organisms observed should be evaluated for size, shape, and Gram reaction, which should be reported with as much description as possible. Reporting the presence of gram-positive cocci in pairs that resemble S. pneumoniae ( Fig. 57.1 ) is more helpful than simply reporting gram-positive cocci in chains. White cells and squamous epithelial cells should be quantified and reported, along with any intracellular bacteria observed. Correlation of Gram stain observations with culture results is a good way to check on the quality of the stains and culture. Demonstration of many bacteria on Gram stain that do not grow out in culture may indicate unusual organisms that require more specialized media or culture conditions. Alternatively, it could suggest a false-positive Gram stain result caused by contamination of reagents or incorrect interpretation of Gram stain results. Gram stain results could indicate the need to inoculate additional media for a specific specimen. For example, finding many gram-negative coccobacilli in a respiratory specimen could indicate the need for a chocolate plate to recover Haemophilus spp., which would not be recovered on a blood agar plate. Other stains, such as the acridine orange stain, can be utilized for staining blood culture bottles, CSF, or buffy coat preparations. This fluorescent stain provides a rapid and, at times, more sensitive stain for bacteria and fungi ( ; ). Bacteria and fungi will produce an orange fluorescence, and mammalian cells will stain green. Some experience is required for accurate interpretation of the acridine orange stain, and correct preparation of the smears is necessary to avoid excessive cellular material, which can result in too much cellular deoxyribonucleic acid (DNA) that can mask the presence of any bacterial DNA.
Rapid recognition of positive blood cultures and timely, appropriate therapy is imperative for improved patient outcomes for sepsis ( ; ). Therefore, rapid identification of pathogens growing in positive blood culture bottles—and, in some cases, identification of markers of resistance—is becoming more common in clinical laboratories. Methods to do this include matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF), in situ hybridization, DNA-microarray hybridization, and nucleic acid amplification from positive blood culture bottles ( ). Detection from patient blood samples without the need for incubation can also be accomplished using magnetic resonance technology ( ). These assays vary in how many pathogens can be detected and what susceptibility or resistance testing is performed.
Media for culture are selected to provide the optimal conditions for growth of pathogens commonly encountered at a particular site or in a particular type of specimen. Consideration is given to special growth requirements of bacteria associated with a given type of infection or to the necessity of selecting certain pathogenic bacteria from a mixed population of usual flora. Therefore, the media chosen may include selective and differential media in addition to standard enrichment agar.
Blood-supplemented agar is a good general growth medium that can be used to demonstrate the hemolytic action of colonies on the red blood cells. Antibiotics or chemicals can be added to create a selective medium, such as colistin–nalidixic acid (CNA) agar or phenylethyl alcohol agar (PEA), both of which are used to inhibit the growth of gram-negative bacilli while permitting gram-positive bacteria to grow. Heating the blood to make chocolate agar and adding vitamin supplements creates an enriched medium with available hemin (X factor) and nicotinamide adenine dinucleotide (V factor) for the isolation of Haemophilus spp. and other fastidious bacteria. Gram-negative bacilli may be separated from gram-positive bacilli by using bile salts and dye in a medium such as MacConkey agar, which additionally divides the colonies into lactose-positive and lactose-negative colonies, thus making it both selective and differential. Guidelines for the selection of media to be used with different types of specimens can be found in multiple references ( ; ).
Bacterial cultures are generally incubated at 35°C. Addition of 5% to 10% carbon dioxide (CO 2 ) may be essential or stimulatory to the growth of N. gonorrhoeae , Haemophilus influenzae , and S. pneumoniae , and should be used whenever feasible. Exceptions to this recommendation are those cultures on differential and selective media in which the pH alteration (which can be affected by added CO 2 ) is used to differentiate colony types (e.g., xylose-lysine-deoxycholate [XLD] agar, Hektoen enteric [HE] agar).
For recovery of anaerobes, inoculated media should be placed into an anaerobic environment as quickly as possible. Several types of anaerobic culture systems are available. One of these is the anaerobic jar, in which water is added to a CO 2 and hydrogen (H 2 ) generator package, and oxygen (O 2 ) is catalytically converted to water with palladium-coated alumina pellets contained in a lid chamber. A modification of this system is a transparent plastic bag containing its own gas generator and palladium catalyst and designed to hold an agar plate; a version of this is the anaerobic Bio-Bag. Another jar-based system uses an automated evacuation-replacement technique to control the anaerobic environment ( ; ).
Another approach to anaerobic culture is the anaerobic glove box or chamber, which consists of a large, clear plastic, airtight chamber filled with an oxygen-free gas mixture of nitrogen, hydrogen, and carbon dioxide. Specimens, plates, and tubes are introduced into or removed from the chamber through a gas interchange lock. Anaerobiosis is maintained by palladium catalysts and the hydrogen gas in the chamber. All manipulations within the chamber are done with neoprene gloves sealed to the chamber wall or, for “gloveless” systems, through a hole with sleeves that seal tightly around the forearms. The chambers contain internal incubators that maintain the incubation temperature. Each of the anaerobe systems has its advantages and disadvantages, but all are equally effective in isolating clinically significant anaerobic bacteria from specimens.
Bacterial cultures should be examined routinely after 18 to 24 hours of incubation. The exception to this is the anaerobe culture, which is generally examined at 48 hours to allow these slower-growing bacteria to produce visible colonies. In general, solid media are held for 48 hours, with liquid media held for an additional 24 to 48 hours. Anaerobic media are incubated for at least 5 days. If this is different for specific organisms, it will be mentioned in the text.
A preliminary report is issued when the culture is first examined; this report is updated as additional information becomes available. Certain results (e.g., positive blood or CSF Gram stain, isolation of an organism requiring infection control measures) are reported to the health care provider as soon as the information becomes available. Final reports are issued when all work on a culture has been completed.
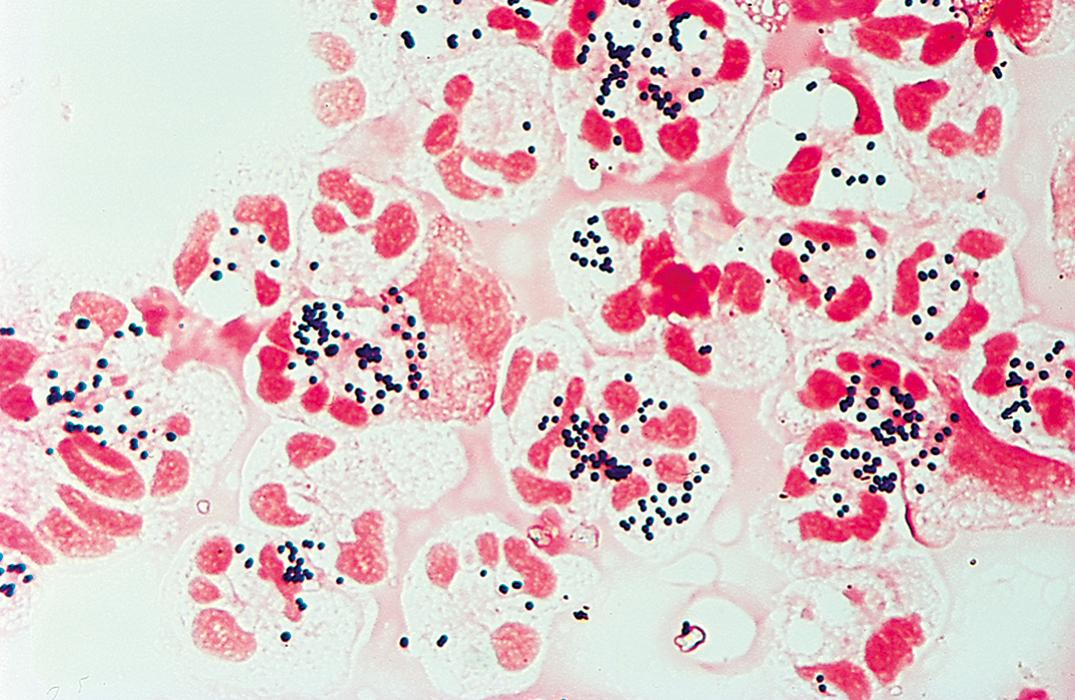
Staphylococci are catalase-positive spherical organisms that often appear in grape-like clusters in stained smears ( Fig. 57.2 ). They grow well on any peptone-containing nutrient medium under aerobic and anaerobic conditions and may produce hemolysis of various species of animal blood cells and yellow or orange pigment on certain types of agar. Growth of staphylococci is readily detected on blood agar plates or in various types of nutrient broth. A selective medium for the isolation of S. aureus is one containing 7.5% to 10% sodium chloride (NaCl) with mannitol ( ).
S. aureus is differentiated from other species of staphylococci principally by its production of coagulases, which are capable of clotting plasma. In addition, many laboratories are now utilizing polymerase chain reaction (PCR) assays for detection of S. aureus in nasal swabs and directly from positive blood culture bottles. Many of these assays enable detection of methicillin-resistant S. aureus (MRSA) versus methicillin-susceptible S. aureus .
S. aureus may be present among the usual flora of the skin, eye, upper respiratory tract, gastrointestinal tract, urethra, and, infrequently, vagina. Therefore, infection may arise from an endogenous or an exogenous source. Factors of importance in the development of infection due to S. aureus include breaks in the continuity and integrity of mucosal and cutaneous surfaces, the presence of foreign bodies or implants, prior viral diseases, antecedent antimicrobial therapy, and underlying diseases with defects in cellular or humoral immunity.
Infections caused by S. aureus may affect multiple organ systems. Among the most common are those involving the skin and its appendages, such as impetigo, folliculitis, mastitis, and infection of surgical wounds. S. aureus is among the leading causes of bacteremia in hospitalized patients, and it may cause endocarditis, particularly in persons with left-sided valvular heart disease and in intravenous drug users. S. aureus is the most common cause of spinal epidural abscess and suppurative intracranial phlebitis, and it may be recovered from brain abscesses, typically following trauma. Meningitis caused by S. aureus is uncommon; it generally follows head trauma or a neurosurgical procedure.
S. aureus is responsible for many cases of osteomyelitis and is the most common cause of septic arthritis. S. aureus is an infrequent cause of community-acquired pneumonia but is a common cause of nosocomial pneumonia, which usually follows aspiration of endogenous nasopharyngeal organisms. Predisposing factors for staphylococcal pneumonia include infection with influenza A virus, cystic fibrosis, and immunodeficiency. Urinary tract infections caused by S. aureus are rare, but cases of pyelonephritis and intrarenal and perirenal abscesses can be found.
Several factors play a role in the virulence of S. aureus . The capsule, if present, has antiphagocytic properties. Cell wall peptidoglycans have endotoxin-like activity, stimulating the release of cytokines by macrophages, activation of complement, and aggregation of platelets. Protein A, an immunologically active substance in the cell wall, has antiphagocytic properties that are based on its ability to bind the Fc fragment of IgG. Other surface proteins, designated as microbial surface components recognizing adhesive matrix molecules, may play an important role in the ability of staphylococci to colonize host tissues ( ). The significant involvement of superantigens of S. aureus in sepsis, endocarditis, and acute kidney injury has been elucidated ( ).
S. aureus produces numerous toxins. The exotoxin TSST-1 is responsible for toxic shock syndrome, and enterotoxins A to E are responsible for staphylococcal food poisoning. The exfoliative toxins—epidermolytic toxins A and B—cause skin erythema and separation, as seen in scalded skin syndrome. Various enzymes are also produced, including protease, lipase, and hyaluronidase, all of which destroy tissue and probably function to facilitate the spread of the infection.
Toxin-mediated diseases caused by S. aureus include scalded skin syndrome, food poisoning, and toxic shock syndrome. Scalded skin syndrome occurs in infants infected with a strain of S. aureus producing exfoliative toxin. The illness begins abruptly with erythema, followed in 2 to 3 days by the formation of flaccid bullae, which slough, leaving denuded areas that eventually resolve completely. Staphylococcal food poisoning, characterized by nausea, vomiting, abdominal cramps, and diarrhea, occurs 1 to 6 hours after ingestion of foods contaminated with preformed staphylococcal enterotoxin, which is usually introduced into the food product by food handlers who prepare and/or serve the food ( ).
Toxic shock syndrome is a multisystem disease affecting individuals who have no antibodies to TSST-1 and are colonized or infected with strains of S. aureus producing TSST-1 or, rarely, enterotoxin B or C. Toxic shock syndrome is primarily the result of a superantigen-mediated cytokine storm and M protein–mediated neutrophil activation, resulting in the release of mediators leading to respiratory failure, vascular leakage, and shock ( ). The illness occurs in women 15 to 25 years of age who use tampons during menstruation, male or female patients with a surgical wound or other focal infection, and individuals who have had a surgical procedure in the nose or sinuses ( ). Toxic shock syndrome begins abruptly with fever, myalgias, vomiting, and diarrhea, followed by hypotension, hypovolemic shock, and an erythematous rash that frequently involves the palms and soles and desquamates in 1 to 2 weeks. The diagnosis is clinical; isolation of S. aureus from any site is not required. Full recovery is the rule, although repeated episodes may occur ( ).
Over the past 10 to 15 years, cases of community-acquired infection with S. aureus that are oxacillin resistant (CA-MRSA) have become more common. In these isolates, Panton-Valentine leukocidin toxin (PVL), which has rarely been associated with hospital-acquired strains of S. aureus ( ), has been found. PVL has been shown to be responsible for necrotizing skin and soft-tissue infections and has been infrequently demonstrated to cause a necrotizing and occasionally fatal pneumonia ( ; ; ; ). Individuals who were initially felt to be at greatest risk are children involved in contact sports and individuals in institutions such as prisons ( ; ). These CA-MRSA strains, unlike hospital-acquired strains of MRSA (HA-MRSA), are often susceptible to most non–β-lactam classes of antibiotics. HA-MRSA strains are usually resistant to all antibiotics except the glycopeptides, such as vancomycin. The mechanism of oxacillin resistance is the same in CA-MRSA and HA-MRSA—the presence of a mecA gene that is responsible for production of a new penicillin-binding protein (PBP-2a or PBP-2′). However, the chromosomal cassette that houses the CA-MRSA mecA gene is different from and much smaller than that containing the mecA gene of HA-MRSA. Many experts believe that, in time, blending of CA-MRSA and HA-MRSA strains will occur, and without molecular typing of any isolate, it will be difficult to distinguish them. Homologs of the mecA gene have been described, including mecC , which results in expression of a PBP2a homolog that has higher affinity for oxacillin than cefoxitin ( ).
S. aureus bacteremia can be caused by community and nosocomial strains of S. aureus . A study compared the risk factors, morbidity, and mortality due to bacteremia related to HA-MRSA bacteremia versus CA-MRSA bacteremia. Patients with CA-MRSA bacteremia were likely to have diabetes mellitus, chronic liver disease, and HIV infection, perhaps reflecting the community nature of acquisition of the S. aureus . In those patients with bacteremia due to HA-MRSA occurring with 48 hours or more of hospitalization, the risk factors were found to include the presence of a central venous catheter, solid tumor, chronic renal failure, prior hospitalizations, and previous antibiotic therapy. Those with bacteremia caused by HA-MRSA that occurred with less than 48 hours of hospitalization were more prone to have had previous hospitalizations, to have been in long-term care facilities, and to have received corticosteroid therapy ( ).
Infections caused by coagulase-negative Staphylococcus (CoNS) spp. usually occur in association with foreign bodies, especially implanted prosthetic valves, joints, and CSF shunts. Isolates of CoNS are usually considered less pathogenic than S. aureus , although that varies among the species and strains ( ). The presence of biofilms and antibiotic resistance of CoNS appears to be associated with bacteremia, whereas specific adhesion capabilities of CoNS were associated with prosthetic joint infections ( ). CoNS are one of the most common organisms associated with CSF shunt infections; they are rarely involved in urinary tract infections, pneumonia, or skin and soft-tissue infections. More than 20 species of CoNS are known, of which S. epidermidis is the species most frequently involved in such infections. S. saprophyticus is an important cause of bacteriuria, particularly among sexually active young women. S. haemolyticus, reported to rank second in frequency to S. epidermidis in clinical specimens, can be resistant to vancomycin, an agent to which most CoNS are susceptible ( ). S. lugdunensis can appear morphologically similar to S. aureus (i.e., in production of a narrow zone of β-hemolysis on blood agar plates) and on occasion will test positive in some assays for coagulase. However, it is usually classified as a coagulase-negative staphylococcus. Clinically, it will act more aggressively than most other CoNS and in this way mimics S. aureus infection, including its role as an agent of endocarditis, osteomyelitis, and other more severe staphylococcal infections ( ). It is important to distinguish S. lugdunensis from other CoNS because the breakpoints to use (according to the Clinical Laboratory and Standards Institute [CLSI]) for the interpretation of susceptibility results versus oxacillin (or cefoxitin) should be those that are used to interpret S. aureus and not those used to interpret breakpoints for oxacillin (or cefoxitin) versus CoNS ( ).
The observation microscopically of gram-positive cocci in clusters in smears of material taken from previously unopened or undrained lesions, or in smears of broth from a positive blood culture, is indicative of staphylococcal infection. All staphylococci are catalase positive. This enzyme converts hydrogen peroxide to oxygen and water, which is detected by observing bubble production when mixing a bacterial colony with hydrogen peroxide.
S. aureus produces coagulase, an enzyme that binds plasma fibrinogen, causing the organisms to agglutinate or plasma to clot. Only rare strains of other staphylococci are coagulase positive. More than 95% of isolates of S. aureus are identified by the slide coagulase test, which detects cell-bound enzyme (clumping factor); nearly 100% of all isolates are identified by tube coagulase tests, which detect free coagulase ( ).
The slide coagulase test is performed by mixing a dense emulsion of the organism with plasma on a glass slide. The test is positive if clumping occurs within 30 seconds. S. lugdunensis and S. schleiferi are two other staphylococci that may give a positive result with this slide coagulase test. A control that consists of emulsifying the suspect colony in saline should be run with each slide test to ensure that autoagglutination does not occur. If autoagglutination is present, slide test results should be considered insufficient for determination of the coagulase reaction of the isolate.
For the tube coagulase test, several colonies are transferred into a tube containing plasma that is incubated at 35°C for 4 hours and then is examined for clot formation. If no clot has formed, the tube is reincubated at room temperature and reexamined after a total of 24 hours of incubation. The test should be examined after 4 hours because most isolates of S. aureus produce a clot within this interval. However, some strains produce a fibrinolysin that can lyse the clot, thus producing a false-negative reaction if the test is observed only after 24 hours. S. intermedius and S. hyicus will also be positive with the tube coagulase test, but they are primarily pathogens of animals and are encountered only rarely in human specimens.
Several commercial latex agglutination assays are available for rapid identification of S. aureus . These assays detect protein A and clumping factor; some also detect capsular polysaccharide, which may improve the ability to detect methicillin (oxacillin)-resistant S. aureus . S. saprophyticus and S. sciuri are two other staphylococcus species that may be latex agglutination positive, along with the rare Micrococcus spp.; however, these should all be slide coagulase negative ( ).
Many species of CoNS have been recognized. However, with the exception of S. epidermidis, S. lugdunensis, and S. saprophyticus , which is resistant to novobiocin, identification of these isolates to the species level is not practical or clinically indicated in every culture. Identification of the species may be needed if isolates are found repeatedly in sterile sites, if susceptibility testing is being performed, if S. lugdunensis is being ruled out, or if correlation between isolates in a patient is being sought to increase the likelihood that the two isolates are the same and, therefore, are potentially clinically relevant. If necessary, attempts to identify these isolates to the species level may be made using commercially available identification kits or with use of MALDI-TOF ( ). Alternatively, isolates may be sent to a reference laboratory capable of performing standard biochemical assays or molecular assays such as 16S ribosomal ribonucleic acid (rRNA) analysis ( ). Staphylococci may be classified into strains for epidemiologic purposes to attempt to identify a common source of infection on the basis of their susceptibility to different bacteriophages, plasmid profiles, cellular fatty acids, electrophoresis of multilocus enzymes, or chromosomal molecular typing (pulsed field gel electrophoresis and repetitive PCR). These tests are generally available only through reference laboratories.
More than 90% of staphylococci are resistant to penicillin due to inducible plasmid-encoded β-lactamase. A penicillin disk diffusion zone edge test or a PCR for the blaZ gene should be performed to confirm a penicillin-susceptible minimum inhibitory concentration (MIC) ( ). Resistance to the penicillinase-resistant penicillins (methicillin, oxacillin, nafcillin) occurs in up to 80% of coagulase-negative staphylococci and in more than 50% of isolates of hospital-acquired S. aureus . Resistance to this group of antimicrobial agents is mediated by the mecA gene, which encodes an altered penicillin-binding protein, PBP2a. Detection of oxacillin resistance is best done by demonstrating resistance to cefoxitin ( ). When MIC testing for oxacillin is performed, the trays should be incubated for 24 hours at 35°C to detect resistance ( ). The mecA homolog, mecC , can be detected by some molecular mecA assays; mecC may be detected in tests for PBP2a ( ).
Several assays have been developed for rapid detection of oxacillin resistance. These include nucleic acid amplification, nucleic acid probe assays for mecA , and latex agglutination assays for PBP2a ( ; ; ; ). For direct detection of MRSA in nasal swabs, two approaches can be used. Chromogenic media specific for the detection of MRSA require overnight incubation but allow easy detection of specific-colored colonies that are positive for the presence of mecA ( ; ; ). Chromogenic agars work well in detecting MRSA but are less sensitive than the available molecular assays. They are, however, less expensive than the molecular assays. Molecular assays that can detect MRSA directly in clinical specimens (nasal swabs, blood cultures, and wounds) are being routinely used in clinical laboratories ( ; ; ; ).
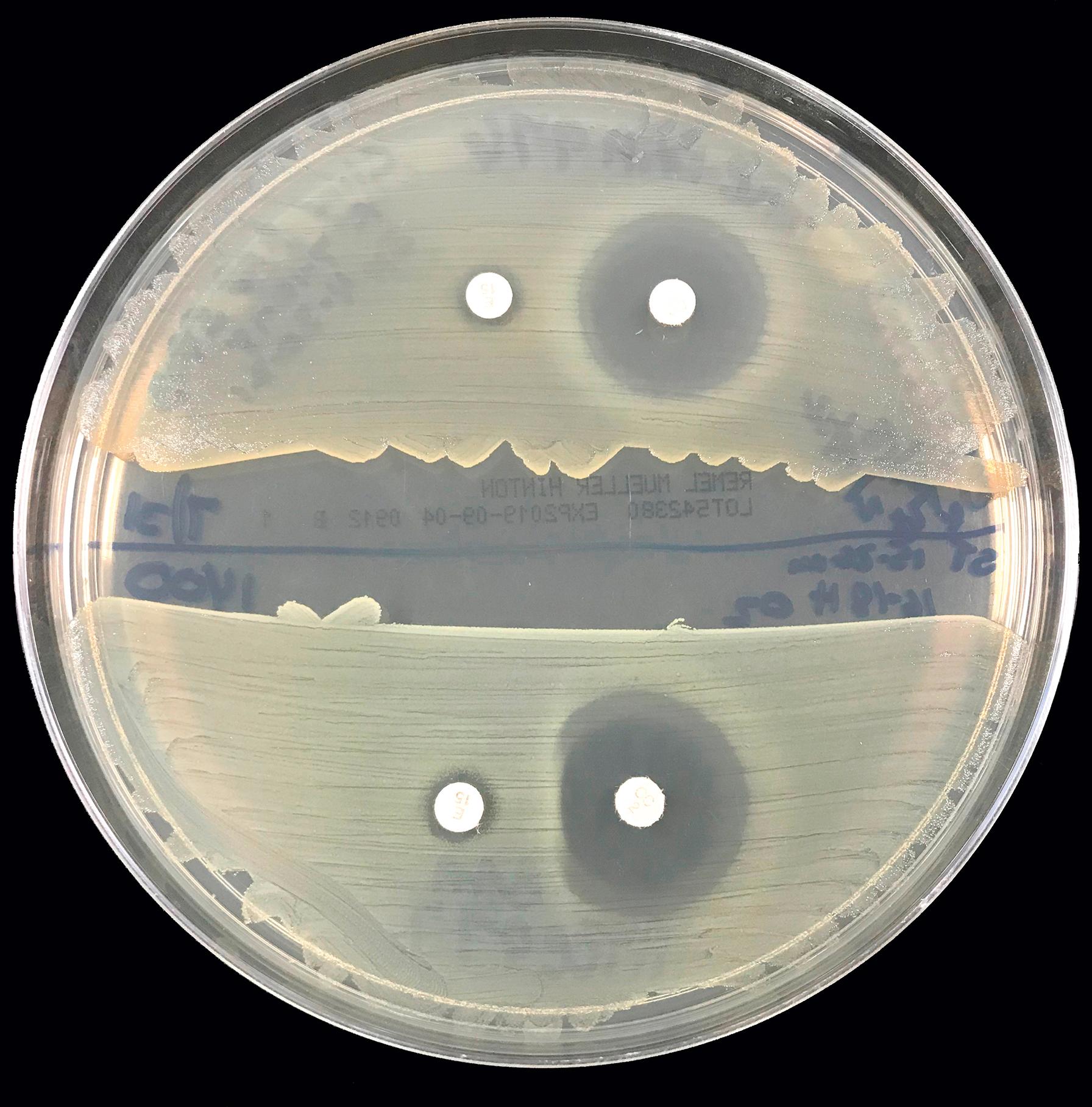
Clindamycin may be used to treat staphylococcal infection. Inducible resistance due to mechanisms involving erm genes may not be detected in routine susceptibility testing. This erm gene also confers cross-resistance to the macrolides (e.g., erythromycin) and streptogramins (quinupristin-dalfopristin). An isolate of S. aureus that is resistant to erythromycin but susceptible to clindamycin should be evaluated for inducible resistance to clindamycin by the “D-zone” test. A 15-μg erythromycin disk and a 2-μg clindamycin disk are placed 15 mm apart on the surface of a blood agar plate inoculated with the isolate in question. After overnight incubation, if there is inducible resistance to clindamycin, blunting of the clindamycin zone of inhibition will be seen on the side near the erythromycin disk, giving the appearance of a D zone ( ) ( Fig. 57.3 ).
Vancomycin resistance, although rarely seen in S. aureus , is a serious issue that laboratories need to be aware of. Isolates of vancomycin intermediately susceptible S. aureus (VISA) have MICs in the intermediate range (4–8 μg/mL) (Cosgrove et al., 2004). In the early 2000s, 14 cases of vancomycin-resistant S. aureus were reported ( ). Because these have not been uniformly detected in automated systems for susceptibility testing, the Centers for Disease Control and Prevention (CDC) recommends that all S. aureus isolates that test resistant to vancomycin be sent to a public health laboratory for confirmation.
Newer antimicrobial agents have good activity against susceptible and resistant staphylococci. These include quinupristin/dalfopristin, a streptogramin; the lipopeptide daptomycin; linezolid; telavancin; and ceftaroline. The last is the only cephalosporin to which MRSA isolates are susceptible.
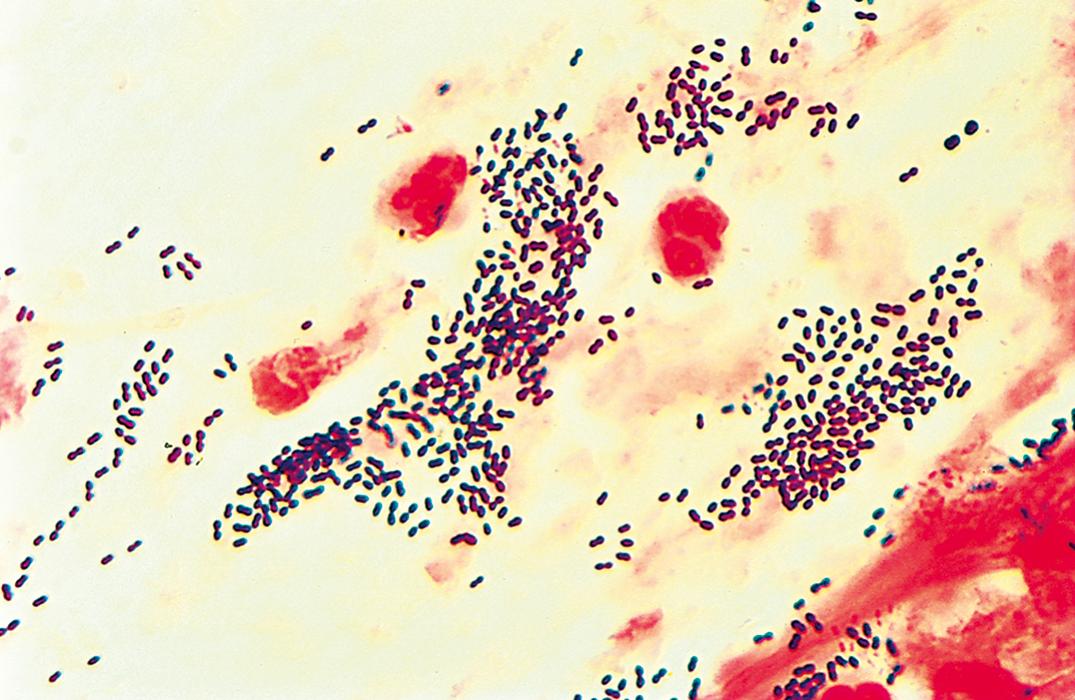
Streptococci are catalase-negative, gram-positive, spherical, ovoid, or lancet-shaped cocci, often seen in pairs or chains. They are facultatively anaerobic. Some strains require added CO 2 for their initial isolation but may lose this requirement in subcultures. Streptococci can be broadly classified according to the hemolytic reaction on blood agar ( Table 57.1 ). Those strains that completely hemolyze the red cells around their colonies are called β-hemolytic and can be further categorized into the Lancefield groups based on serologically reactive carbohydrates. Important members of this group include S. pyogenes (group A) and S. agalactiae (group B). Figure 57.4 is a Gram stain of S. pyogenes (group A streptococcus) in a specimen from abscess material on the arm of a patient with cellulitis. The gram-positive cocci in chains that produce partial hemolysis (which cause “greening” of the agar) are α-hemolytic. An important member of this group is S. pneumoniae . Streptococci that do not hemolyze blood are γ-hemolytic. An important member of this group is the S. bovis complex. Some S. agalactiae may also be γ-hemolytic. Most of the remainder of the α- and γ-hemolytic streptococci are collectively called viridans streptococci, including S. mutans , S. sanguis , S. mitis , S. salivarius , and S. anginosus . The group of organisms previously referred to as nutritionally variant (pyridoxal or thiol dependent, satelliting) streptococci have now been assigned to the genus Abiotrophia or Granulicatella .
| Hemolysis | Lancefield Group | Species |
|---|---|---|
| β | A | Streptococcus pyogenes |
| B | Streptococcus agalactiae | |
| C | Streptococcus dysgalactiae | |
| D | Enterococcus spp. | |
| α or γ | D | Enterococcus spp. |
| D | Streptococcus bovis complex (reclassified into many new species, as described in text) | |
| None | Viridans group ∗ | |
| α | None | Streptococcus pneumoniae |
∗ Small colony variants of Lancefield group A, C, F, or G, or nongroupable strains, can be any hemolysis.
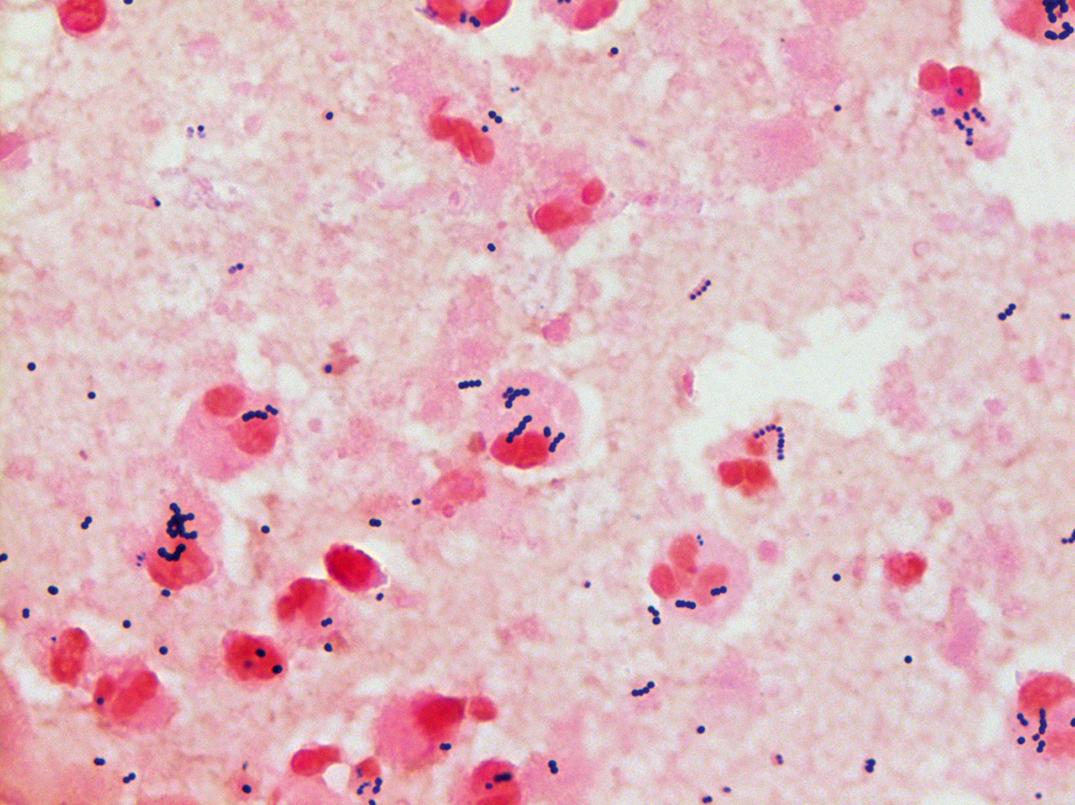
Members of the genus Enterococcus , previously designated as group D streptococci because their cell wall antigens reacted with group D antisera, are different molecularly as well as metabolically from the other members of the genus Streptococcus . These organisms are gram-positive cocci that occur singly, in pairs, and in short chains. They are facultatively anaerobic. Most enterococci are α- or γ-hemolytic on blood agar, but some may exhibit β-hemolysis. The most common species are E. faecium and E. faecalis ; yellow motile strains of the enterococci, less commonly associated with infections, include E. casseliflavus and E. gallinarum . These latter two species are usually intrinsically vancomycin resistant; it is important to differentiate them from the vancomycin-resistant E. faecium or E. faecalis strains.
Other genera of catalase-negative gram-positive cocci that may be isolated from clinical specimens include Leuconostoc , Pediococcus , Rothia , Gemella , Aerococcus , Lactococcus , and other less rarely isolated species. These organisms are considered to have low virulence potential and generally are pathogenic only in the compromised host, with the exception of some species of Aerococcus ( A. urinae and A. sanguinicola ), which are known urinary tract pathogens. However, some of these isolates may be confused with viridans streptococci, in particular; their differentiation is important because of their lower virulence and potential for vancomycin resistance.
One of the most common clinical manifestations of group A streptococci is pharyngitis. This may be accompanied by scarlet fever, a punctate exanthem overlying diffuse erythema that usually first appears on the neck or upper chest, becomes generalized, and then desquamates. Skin infections caused by group A streptococcus include cellulitis, erysipelas, and pyoderma. Acute rheumatic fever, characterized by carditis, polyarthritis, erythema marginatum, chorea, and subcutaneous nodules, may occur 1 to 5 weeks after group A streptococcal pharyngitis. Acute glomerulonephritis may develop 10 days to 3 weeks after group A streptococcal pharyngitis or pyoderma.
Beginning in the late 1980s, serious group A streptococcal clinical syndromes, including necrotizing fasciitis, myositis, malignant scarlet fever, bacteremia, and toxic shock–like syndrome, began to be seen with increasing frequency. These have been associated with high morbidity rates and mortality rates of up to 30% or more. The reason for this increase is not completely understood but appears to be related to changes in the prevalence of organisms having an enhanced virulence potential ( ; ; ; ).
S. pyogenes produces numerous virulence factors. One of the most important is the antiphagocytic cell wall M protein. Antibodies against the specific M protein confer lifelong type-specific immunity. However, because more than 60 M protein types exist, infection with a group A streptococcus possessing a different M protein may occur. Another important cell wall component is lipoteichoic acid, which permits bacterial adherence to the respiratory epithelium. S. pyogenes also elaborates about 20 extracellular products, including enzymes (streptolysins, hyaluronidase, streptokinase, deoxyribonucleases [DNases], and nicotinamide adenine dinucleotidase [NADase]) and erythrogenic toxins. Streptolysin O, an antigenic, oxygen-labile enzyme, produces subsurface hemolysis on blood agar plates; streptolysin S, a nonantigenic, oxygen-stable enzyme, produces surface hemolysis. Neither streptolysin has a proven role in the pathogenesis of human disease. Streptokinase promotes fibrinolytic activity by converting plasminogen to plasmin, and hyaluronidase may enhance the spread of the organism through connective tissue. The pathogenic significance of the DNases and of NADase is unknown. Pyrogenic (erythrogenic) toxins (serotypes A, B, C) are produced by isolates of S. pyogenes infected with a specific temperate bacteriophage. Their pyrogenicity is caused by a direct action on the hypothalamus. Streptococcus group A has also been found to possess superantigens with high mitogenic capabilities. They have been associated with cases of more severe streptococcal infection, such as necrotizing fasciitis or toxic shock syndrome ( ; ).
The pathogenesis of acute rheumatic fever is not fully understood. Certain M protein types of S. pyogenes may be rheumatogenic. The presence of complexes of immunoglobulin and the C3 component of complement along the sarcolemmal sheaths of cardiac myofibers from individuals with rheumatic carditis suggests that myocarditis results from the production of antibodies directed against a streptococcal cell wall M protein that cross-reacts with myocardial tissue. Moreover, a heart or tissue cross-reactive antigen of S. pyogenes that shares immunologic epitopes with, but is distinct from, the M protein has been identified ( ). Renal damage in acute glomerulonephritis is caused by deposits of circulating streptococcal–antistreptococcal immune complexes in the glomeruli and subsequent activation of complement. Cell-mediated reactions to an altered glomerular basement membrane or activation of the alternate complement pathway also may be involved. Isolates of S. pyogenes have been linked to toxic shock syndrome, with a clinical picture very similar to S. aureus ; streptococcal superantigens (Sags) are thought to be responsible for the toxic shock–like syndrome caused by strains of S. pyogenes ( ).
The most significant infections caused by Group B streptococci (GBS) are neonatal sepsis, pneumonia, and meningitis. Colonization of the maternal genital tract is associated with colonization of infants and risk of neonatal disease, with early-onset infections occurring within the first few days after delivery and late-onset infections appearing after 1 week of age. To reduce the incidence of neonatal disease, the CDC published specific guidelines to facilitate early identification and treatment of women colonized with GBS and identification and treatment of neonates at risk for developing disease (ACOG, 2019; Verani et al., 2010). All pregnant women at 36 0/7 to 37 6/7 weeks of gestation should have vaginal/rectal specimens collected and processed for detection of GBS. These specimens can be cultured or have a nucleic acid amplification test (NAAT) performed on them after enrichment in a selective culture broth (ACOG, 2019). Results of this test should be available during labor so that appropriate prophylaxis can be given to the mother before delivery to prevent infection of the newborn. Isolation of >10 4 colony-forming units (CFUs) per milliliter of GBS from the urine of a pregnant female can also be used as a marker of GBS vaginal carriage. This information should be used to direct prophylaxis to mothers found to be positive. If urine is positive, screening of vaginal/rectal cultures may not be necessary. Molecular tests that can rapidly identify the presence of GBS during or right before delivery are available, although reported sensitivities vary and susceptibility testing for clindamycin in penicillin-allergic patients is not possible (ACOG, 2019; ). GBS infections in adults include postpartum endometritis, urinary tract infection, bacteremia, skin and soft-tissue infections, pneumonia, endocarditis, meningitis, arthritis, and osteomyelitis.
Group C and G streptococci are similar to S. pyogenes in that they cause a wide range of infections, including bacteremia, endocarditis, meningitis, arthritis, and respiratory and skin infections ( ). The pharyngeal infection caused by these streptococci is similar to that of group A streptococci except that the nonsuppurative sequelae of rheumatic fever do not occur.
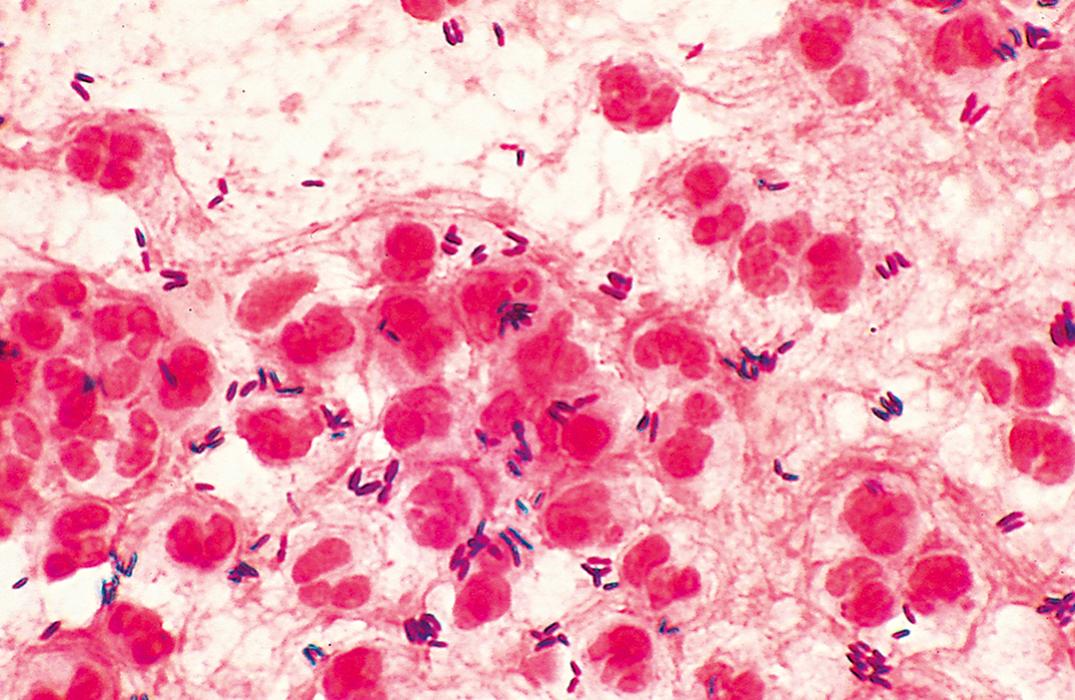
Infections caused by S. pneumoniae include pneumonia, meningitis (especially in infants and the elderly), spontaneous bacteremia (in persons who do not have a spleen), otitis, sinusitis, and spontaneous peritonitis. S. pneumoniae is seen in the normal flora of the upper respiratory tract of 25% to 50% of preschool children, 36% of primary school–age children, and nearly 20% of adults, termed carriers with decreasing prevalence of invasive serotypes due to vaccination ( ; ). Its spread is enhanced by upper respiratory tract infections and crowding. Pneumonia may develop when the host immune defenses are impaired. Most cases are endogenous, following aspiration of oral secretions containing normal flora that includes S. pneumoniae . Person-to-person transmission during epidemics occurs by droplet aerosols. The major virulence factor of S. pneumoniae is its antiphagocytic polysaccharide capsule; strains with a thick, mucoid capsule are especially virulent. Vaccines designed to protect against infection by pneumococci of many of the predominant capsular polysaccharide types are available. The CDC recommends that infants receive the 13-valent conjugated vaccine starting at 2 months of age. It also is recommended that adults 65 years of age and older receive the 23-valent polysaccharide vaccine. Immunosuppressed patients of any age should receive both vaccines ( ).
Bacterial endocarditis is the most common infection caused by viridans streptococci; others include abscesses in the brain or liver, bacteremia, and dental caries. The Streptococcus anginosus group ( S. constellatus , S. intermedius , and S. anginosus ) consists of the most common viridans streptococci responsible for liver, spleen, and brain abscesses. They often are more susceptible to antibiotics than other strains of viridans streptococci, although resistance to penicillin is increasingly being recognized. S. bovis bacteremia has been associated with malignancies of the gastrointestinal tract. S. bovis is now recognized as a complex of seven subspecies broken down into four branches: S. gallolyticus (includes the subspecies gallolyticus , pasteurianus , and macedonicus ); S. equinus ; S. infantarius (includes subspecies infantarius and coli ); and S. alactolyticus . S. gallolyticus subsp. gallolyticus is isolated from blood cultures of patients with colonic cancer more often than others in the group . S. gallolyticus subsp. gallolyticus is associated with prosthetic joint infections and infective endocarditis; S. gallolyticus subsp. pasteurianus is associated with meningitis in infants. S. alactolyticus , S. equinus , and S. infantarius are not often found in human disease ( ; ). In many laboratories, isolates are still reported as S. bovis because phenotypic identification may not be adequate for differentiation and physicians recognize the clinical significance of S. bovis . Other viridans group streptococci include the Streptococcus mitis group (commonly associated with endocarditis), the Streptococcus mutans group (etiologic agent of dental carries), and the Streptococcus salivarius group (isolated from the oral cavity and blood). As use of molecular methods or MALDI-TOF for bacterial identification increases, more of these differences may be appreciated ( ; ).
Enterococci are not highly pathogenic; however, they are a common cause of urinary tract infection in hospitalized persons. They may also cause endocarditis, bacteremia, and wound infections. Vancomycin-resistant enterococci offer a greater potential for infection, especially in immunocompromised patients and patients with implanted foreign devices ( ; ).
Streptococci grow well on blood or chocolate agar. Blood agar is preferred because the hemolytic properties of the organism can be assessed. When culturing vaginal/rectal swabs from pregnant women for GBS, specimens should first be inoculated to a selective broth, such as Lim or carrot broth, or on to selective agar, such as Granada agar, to enrich for this organism ( ; ; ). Tests that may be used in the clinical microbiology laboratory to presumptively name the β-hemolytic species of Streptococcus are shown in Figure 57.5 . More than 99% of isolates of group A Streptococcus (GAS) are susceptible to bacitracin, but a very small percentage of isolates of GBS and 10% to 20% of isolates of groups C and G Streptococcus are also susceptible. Therefore, results of the bacitracin susceptibility test provide a presumptive identification. An isolate may be called GAS presumptively based on hydrolysis of l -pyrrolidonyl-β-naphthylamide (PYR) ( ). All isolates of GAS and more than 99% of isolates of Enterococcus are PYR positive. Identification of GAS is confirmed by bacitracin susceptibility or using latex agglutination for the Lancefield group A antigen.
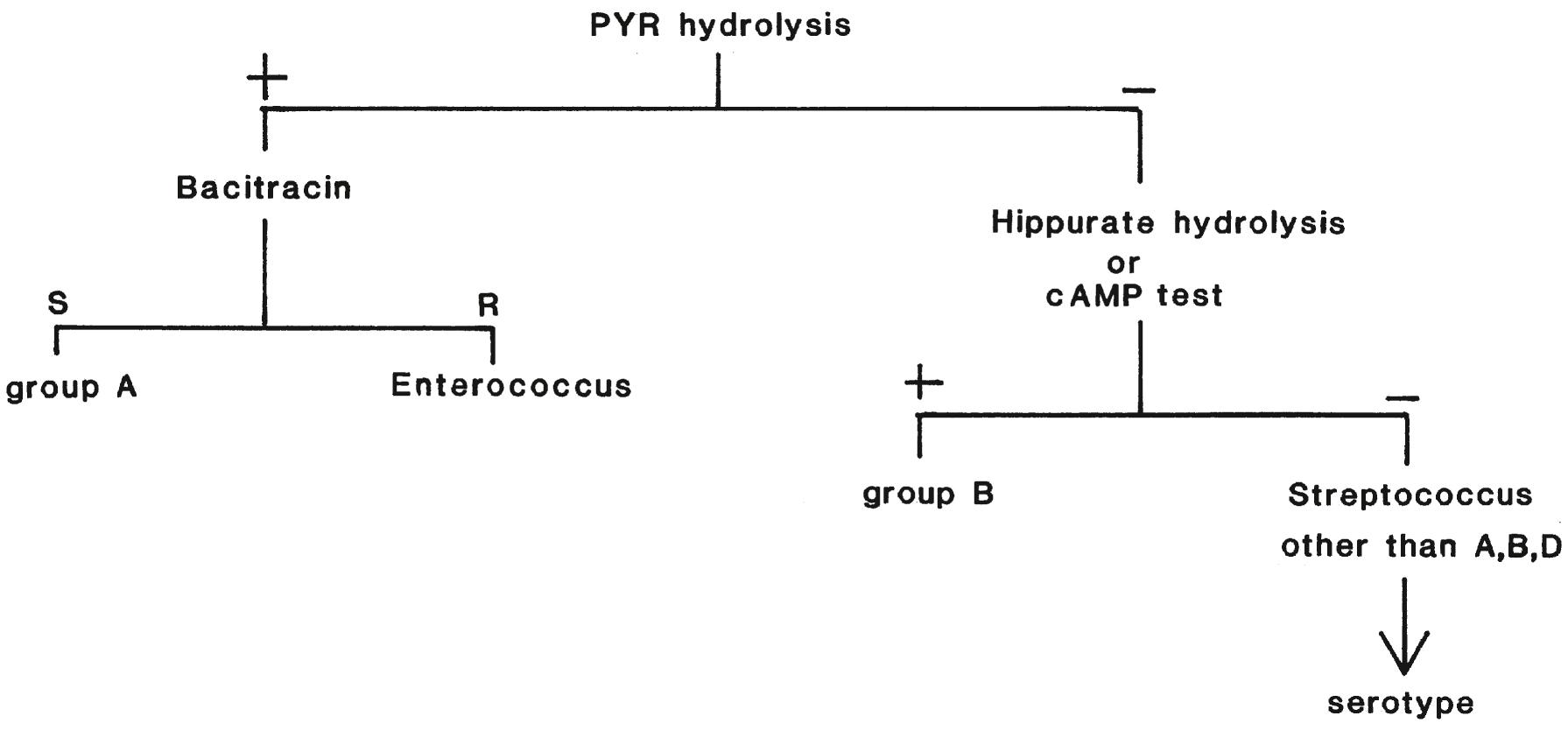
GAS antigen may be detected directly in throat swab specimens by using commercial kits designed to generate a rapid result. These tests are highly specific but, given their low sensitivity, which varies among studies from 31% to 95% ( ; ), a negative antigen test in children should be followed by culture or NAAT. More recently, clinical guidelines recommended that negative antigen assays in adults need not be followed up with culture ( ; ; ; ). The use of NAAT for direct diagnosis of GAS pharyngitis can allow laboratories to perform single-tiered testing without reflex cultures ( ). Serologic tests to detect streptolysin O and DNase B antibodies in acute and convalescent serum samples are used primarily to diagnose acute rheumatic fever and acute glomerulonephritis following infection with GAS.
Catalase-negative colonies that are β-hemolytic and hippurate hydrolysis positive and/or that have a positive CAMP test (named for researchers Christie, Atkins, and Munch-Petersen) reaction presumptively can be called group B Streptococcus (GBS). Isolates of presumed GBS from sterile body sites should be identified by serotyping (using latex agglutination or coagglutination tests) or MALDI-TOF. For culture of vaginal/rectal swab specimens from pregnant women during weeks 36 through 38 of gestation, it is recommended that a broth enrichment be used along with or as a replacement for agar-based media. Selective broth media, including Lim broth or selective Todd-Hewitt broth, can be used as enrichment media ( ). Chromogenic broth, including carrot broth media, can be used as enrichment broth; colonies of β-hemolytic GBS will convert the color of the tube from clear to yellow or orange. However, nonhemolytic GBS will not change the tube color and, when used, a negative broth would still need to be planted onto solid media for recovery of these strains. In addition, a selective nonchromogenic enrichment broth can be subcultured to Granada agar, on which colonies of GBS will appear yellow to orange for ease of detection. Nucleic acid amplification assays can be used to detect GBS directly in vaginal-rectal specimens or can be used to detect GBS in Lim or carrot broth cultures ( ; ; Gray et al., 2012). Current guidelines highlight the higher sensitivity of NAAT performed after enrichment culture (MMWR, 2010).
Latex agglutination assays are available for direct detection of GBS (as well as S. pneumoniae , some serotypes of N. meningitidis , E. coli , and H. influenzae type b) in CSF, serum, and urine. These assays have been shown to have sensitivities equivalent to or lower than a Gram stain and may give false-positive results. The rapid bacterial antigen tests are much more expensive and labor-intensive than Gram stain, so most laboratories no longer offer these tests or strictly limit their use ( ). Nucleic acid amplification tests for detection of common pathogens in central nervous system infections have been developed and can decrease the time to detection of pathogens relative to cultures. These assays can have false-positive and false-negative results; thus, discussions with health care providers on how to best utilize this test are required ( ).
Tests used to presumptively identify α- and γ-hemolytic streptococci and enterococci are shown in Figure 57.6 . α-Hemolytic colonies that are mucoid or flattened with a depressed center are suggestive of S. pneumoniae ; they should be tested for susceptibility to optochin (P disk), and for bile solubility. S. pneumoniae is susceptible to both; other α-hemolytic streptococci are resistant to optochin and are variable in response to bile. A urinary antigen assay for detection of S. pneumoniae has been shown in some studies to be a nonculture diagnostic method for patients with severe pneumococcal infection, for diagnosis of pneumococcal exacerbation in chronic obstructive pulmonary disease patients, and as a tool for diagnosis of otitis media. Reported sensitivities compared with conventional diagnostic methods range between 50% and 80% with specificities of at least 90% ( ). As with any antigen assay, caution needs to be taken in interpreting results in cases in which prior infection with S. pneumoniae may have occurred since the assay can remain positive for 1 to 6 months ( ; ; ; ).
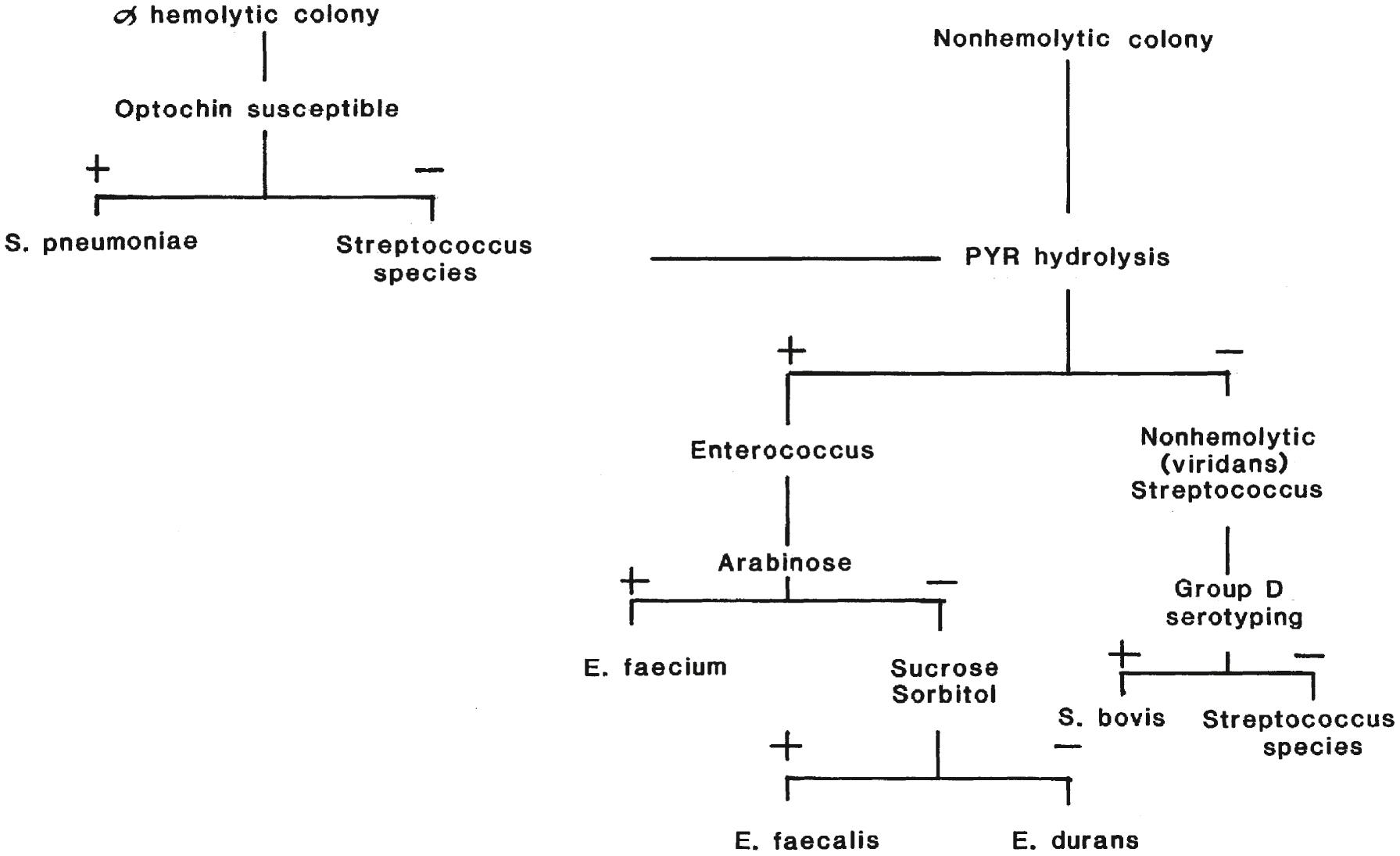
α-Hemolytic colonies that are not S. pneumoniae and γ-hemolytic colonies are tested for PYR hydrolysis; enterococci are PYR positive, and viridans streptococci are negative. Moreover, all enterococci grow in the presence of 6.5% NaCl, but viridans streptococci do not. Enterococci hydrolyze esculin in the presence of bile (causing visible growth and blackening of the agar), but up to 10% of viridans streptococci are also bile–esculin positive. Additional biochemical assays or MALDI-TOF are required to identify enterococci to the species level. A majority of vancomycin-resistant enterococci (VRE) are E. faecium .
α-Hemolytic and γ-hemolytic streptococci that are not S. pneumoniae or Enterococcus are grouped as viridans streptococci. Identification of individual species of viridans streptococci requires conventional biochemical testing, molecular methods, or MALDI-TOF ( ; ). Kit systems to identify these organisms are commercially available although they may lack accuracy for viridans group streptococci. Full identification of species members of viridans streptococci, however, is usually not necessary.
Leuconostoc and Pediococcus rarely cause disease, but it is important to correctly identify them as they are intrinsically resistant to vancomycin. Characteristics that can be used to identify and differentiate these organisms are listed in Table 57.2 ( ). Included in this table is Aerococcus spp., because of their morphologic similarity to Enterococcus spp. The clue that an isolate (especially from a urine culture) might be an Aerococcus sp. is the finding on Gram stain that a catalase-negative colony consists of gram-positive cocci in tetrads and clusters, not in pairs and chains. Some species of Aerococcus are PYR positive, which leads to further confusion with Enterococcus spp.
| Enterococcus | Aerococcus | Leuconostoc | Pediococcus | |
|---|---|---|---|---|
| Gram stain | Pairs and short chains | Tetrads | Cocci, coccobacilli, and rods; pairs and chains | Tetrads and pairs; spherical cells |
| Hemolysis | β or α or γ | α or γ | α or γ | α or γ |
| Bile esculin | + | V | V | + |
| Growth in 6.5% NaCl | + | + | V | V |
| PYR | + | ∗ | – | – |
| LAP | + | ∗ | – | + |
| Vancomycin susceptibility | S/R | S | R | R |
∗ Aerococcus urinae is PYR and LAP positive; Aerococcus viridans is PYR positive and LAP negative.
Members of the genus Abiotrophia and Granulicatella will not grow in the absence of pyridoxal. Often, they are first recognized as satelliting colonies growing around a colony of S. aureus or only growing on chocolate media ( ). Methods of performing susceptibility tests for strains of Abiotrophia and Granulicatella can be found in the third edition of CLSI document M45 ( ).
Groups A, B, C, and G streptococcus are all are susceptible to penicillin. Therefore, routine antimicrobial susceptibility testing of these organisms is unnecessary unless penicillin cannot be used, as in the case of a penicillin allergy. In those situations, testing for resistance to macrolides, clindamycin, and the tetracyclines may be warranted. Inducible resistance to clindamycin may occur with Streptococcus (i.e., GBS), and a D zone test as described earlier for S. aureus may be warranted if the streptococcal isolate is found resistant to erythromycin. Specific guidelines for streptococcal D–zone testing recommend that the clindamycin and erythromycin disks be separated by 12 mm instead of the 15 to 16 mm recommended for testing S. aureus ( ).
Because S. pneumoniae organisms with intermediate- or high-level resistance to penicillin are found worldwide, isolates should be tested for susceptibility to penicillin. A screening test using disk diffusion with a 1-μg oxacillin disk (≥20 mm = susceptible) may be performed. However, isolates that are not susceptible by this method must be further evaluated by macrodilution or microdilution testing using Mueller-Hinton broth supplemented with lysed horse blood or gradient diffusion testing to determine the penicillin MIC. There are breakpoints for penicillin and the cephalosporins for isolates from cases of meningitis versus those isolated from nonmeningitis sites ( ). Resistance to third-generation cephalosporins also occurs; therefore, isolates should be tested for susceptibility to these antimicrobial agents as well.
Susceptibility testing should be performed on isolates of nonhemolytic (viridans) streptococci from sterile body sites, because resistance to penicillin does occur. Enterococcus spp. should also be tested, primarily to identify high-level resistance to penicillin or ampicillin, high-level resistance to streptomycin and gentamicin, and resistance to vancomycin. Enterococci are resistant to vancomycin because of the presence of resistance genes, referred to as the van genes ( ). Although many of these genes have been described, the most common are vanA , vanB , and vanC . The vanA and vanB genes, conferring high-level resistance and predominantly found in E. faecium and less frequently in E. faecalis , are acquired, plasmid-borne genes that can create infection control problems involving transmission of this resistance. This vancomycin resistance should be differentiated from intrinsic and lower-level resistance ( vanC genes) in the yellow, motile species of Enterococcus ; this information should be reported to the infection control team ( ).
Other nonstreptococcal gram-positive, catalase-negative cocci of increasing importance are those that belong to the genera Gemella and Aerococcus . Gemella spp. ( G. haemolysans and G. morbillorum ) resemble viridans streptococci, although they usually produce smaller colonies. G. haemolysans has been associated with endocarditis and meningitis. Gram stain usually demonstrates diplococci with adjacent sides flattened that can be confused with a Neisseria sp. because cells can easily become decolorized. G. haemolysans prefers an aerobic growth atmosphere. G. morbillorum usually appears as cocci in pairs or short chains. Both are PYR positive, 6% NaCl, and esculin–hydrolysis negative (to differentiate them from enterococci). G. morbillorum is leucine-aminopeptidase (LAP) positive as well. Both are usually susceptible to penicillin. As with other gram-positive cocci, if there is doubt about the morphology of the organism (i.e., whether it is a short rod or a coccus), Gram stain performed from a broth culture will usually resolve the difficulty. MALDI-TOF has been shown to be a useful tool in the correct identification of Gemella spp. ( ).
Two major species of Aerococcus may be clinically relevant and/or isolated from clinical specimens: A. urinae and A. viridans . Both resemble viridans streptococci or enterococci on agar plates. However, in Gram stains, they usually appear in tetrads. An increasing number of clinically relevant cases of A. urinae (and other species of aerococci) are being reported. Urinary tract infections are most common, but rare cases of more serious infection, including bacteremia, are seen ( ; ). A. viridans remains relatively uncommon as a pathogen when isolated from clinical samples; A. sanguinicola isolation is not always associated with clinical disease, but case numbers are increasing ( ; ).
A. urinae is PYR negative and LAP positive, in contrast to A. viridans , which is PYR positive and LAP negative (see Table 57.2 ). Both will grow in the presence of 6.5% NaCl. Neither are anaerobes, and A. viridans usually will not grow under anaerobic conditions. A. urinae is usually susceptible to penicillin and nitrofurantoin but may be resistant to sulfonamides. Variability in its response to trimethoprim has been noted ( ; ). A newer member of the genus Aerococcus , A. sanguinicola , which is rarely recovered from clinical specimens, can be both LAP and PYR positive, although this is not a confirmatory identification. Use of MADLI-TOF can provide more reliable results. It can be responsible for urinary tract infections as well as bacteremia and infective endocarditis. Like A. urinae , isolates of A. sanguinicola are usually susceptible to penicillin and nitrofurantoin, but are variable in their response to sulfonamides and quinolones ( ; ; ).
The corynebacteria, also known as diphtheroids , appear in the gram-stained smear as slightly curved, gram-positive rods with nonparallel sides and sometimes wider ends, giving a clubbed appearance ( Fig. 57.7 ). These organisms are catalase positive. More than 100 species of Corynebacterium are known. Most are rarely pathogenic in humans; notable exceptions are C. diphtheriae and its closely related species C. ulcerans and C. pseudotuberculosis . C. pseudodiphtheriticum has been implicated in respiratory tract infections, including pneumonia ( ). Other medically relevant coryneform gram-positive rods include Arcanobacterium haemolyticum , Trueperella pyogenes , and Trueperella bernardiae . These organisms also appear as irregular gram-positive rods on Gram stain but can be easily differentiated from the corynebacteria by their negative catalase reaction.
At the initial site of infection on the epithelial cells of the tonsils and oropharynx, C. diphtheriae elaborates an exotoxin that causes local cell necrosis and subsequent inflammation. The exotoxin is encoded by a specific bacteriophage carrying the tox gene and is absorbed into the circulation. Distribution of exotoxin through the bloodstream can produce degenerative changes in the heart, nervous system, and kidneys. The toxin molecule consists of two fragments: A, containing the enzymatically active site, and B, comprising the receptor binding site. Once in the cell, protein synthesis is disrupted. The bacteria and exotoxin produce a serum exudate and cellular infiltrate of the mucous membrane in the pharynx. Exudative lesions coalesce, forming a grayish-black adherent pseudomembrane, which is characteristic of diphtheria. Although toxin production and pathogenicity are often considered to be synonymous, pseudomembranes may form in persons infected with nontoxigenic strains. Extension of the pseudomembrane superiorly into the nasopharynx or inferiorly into the larynx may be so marked as to produce respiratory obstruction. Although C. diphtheriae infections of other parts of the body do occur, those observed most frequently in the United States today are infections of the skin. Transmission of C. diphtheriae occurs by droplet nuclei from the respiratory tract or by contact from cutaneous foci of infection ( ; ). Nontoxigenic strains of C. diphtheriae can cause skin and bloodstream infections ( ).
Because they are part of the normal flora of the skin and mucous membranes, it is difficult to establish the etiologic role of the other corynebacteria. Clinical significance is generally increased if the organism is observed in the gram-stained smear in association with leukocytes, is isolated from a sterile site, and is isolated from multiple samples. C. jeikeium has been clearly associated with infections of implanted prosthetic materials (e.g., heart valves, CSF, joints), has caused subacute bacterial endocarditis, and has been involved in a variety of opportunistic infections. C. urealyticum has been associated with urinary tract infection as well as with bacteremia, endocarditis, and wound infection ( ). When identified, C. striatum and C. amycolatum are the most common normal flora skin coryneforms. They may become pathogenic, especially in cases of prosthetic joint infections and, of note, are often resistant to β-lactam antibiotics, a characteristic usually attributed only to C. jeikeium ( ; ).
A. haemolyticum has been associated with pharyngitis and wound and soft-tissue infections ( ). T. pyogenes and T. bernardiae are associated with abscess formation ( ).
Because of the relative rarity of diphtheria in the United States today, the diagnosis may be overlooked clinically, and the laboratory may easily fail to recognize it in cultures. When the diagnosis of diphtheria is suspected, the laboratory should be informed so that the specimen can be handled appropriately. Specimens should be obtained with a cotton- or polyester-tipped swab from inflamed regions of the nasopharynx and, if possible, beneath the pseudomembrane. If skin lesions are suspected of being positive for C. diphtheriae , the most appropriate specimen would be an aspirate of the lesion. Corynebacteria will grow on routine blood-containing agar; however, cystine-tellurite (CT) blood agar or Tinsdale medium is preferred. On CT medium, colonies of C. diphtheriae are gray or black after 48 hours of incubation. Colonies may be large or small and flat or convex. Colonies of species other than C. diphtheriae may produce black colonies on CT or Tinsdale media, although these will usually be smaller. If a laboratory does not have CT or Tinsdale medium and a request for C. diphtheriae is made, CNA can be used, although it will be more difficult to recognize possible C. diphtheriae strains ( ).
Classification of oral and skin corynebacteria or diphtheroids is difficult and confusing. Commercial identification systems, including MALDI-TOF, are available to identify many of the members of this group of organisms ( ). Isolates of suspected C. diphtheriae must be tested for production of exotoxin. The elaboration of toxin may be detected in vitro with the Elek immunodiffusion test. However, this generally is not done in a routine clinical laboratory. Isolates should be sent to a public health laboratory or a reference laboratory for an Elek immunodiffusion test. Alternatively, PCR-based tests available at reference or public health laboratories have been described that may be used for detection of the toxin gene ( ). C. jeikeium often produces a characteristic metallic sheen on the surface of blood agar plates. C. striatum and C. amycolatum are common skin flora that can be responsible for infection. Whether they need to be specifically identified is controversial, and identification can be difficult with biochemical methods. Species identification of Corynebacterium often requires a combination of commercial kit systems, cellular fatty acid analysis, MALDI-TOF, and/or sequencing ( ). C. striatum and C. amycolatum organisms are often resistant to many antibiotic agents—a characteristic that often is more typically associated with hospital-acquired strains, in particular, C. jeikeium . MALDI-TOF has been reported to be very useful in the identification of Corynebacterium spp. ( ; ).
Arcanobacterium spp. are β-hemolytic on sheep blood agar. Colonies on sheep blood agar are small after 48 hours of incubation, and the hemolysis may go unnoticed. Adequate growth and noticeable hemolysis are best demonstrated in a CO 2 -enhanced environment. Arcanobacterium spp. are catalase negative. Biochemical reactions that are used to determine the species of corynebacteria are also useful in identifying the Arcanobacterium spp. ( ). A. haemolyticum produces phospholipase D, which is responsible for the reverse CAMP reaction with S. aureus . This organism inhibits hemolysis around the S. aureus streak, producing an inverted triangle of no hemolysis.
Although antitoxin remains the only specific method of treatment of diphtheria, antibiotics are administered to patients with disease and to asymptomatic carriers of toxigenic strains. C. diphtheriae is usually inhibited by penicillins and the macrolides. The antimicrobial susceptibilities of other species of corynebacteria or diphtheroids are far less predictable. C. jeikeium is usually resistant to the penicillins and cephalosporins, is variably susceptible to most other antibiotics, and is almost uniformly susceptible to vancomycin. Other species of Corynebacterium , however, may be similarly resistant to β-lactam antibiotics. Treatment of infection caused by these organisms is often complicated by the presence of compromised host defenses and implanted prosthetic materials. Arcanobacteria are sensitive to penicillin and other β-lactams, rifampin, tetracycline, and the macrolides. Growth of the organisms may be inhibited by fluoroquinolones and aminoglycosides ( ). Methods used for performing susceptibility tests on Corynebacterium spp. and interpretive criteria can be found in CLSI document M45-A3 ( ). Susceptibility patterns of Corynebacterium spp. have been recently described ( ).
Methods of prevention of diphtheria are almost exclusively active and passive immunization programs with supplemental antibiotics to eliminate the carrier state of toxigenic strains during epidemics. Vaccination for diphtheria is with a modified version of the toxin (a toxoid). Thus, colonization or infection with a no-toxigenic strain of C. diphtheriae is still possible in vaccinated individuals.
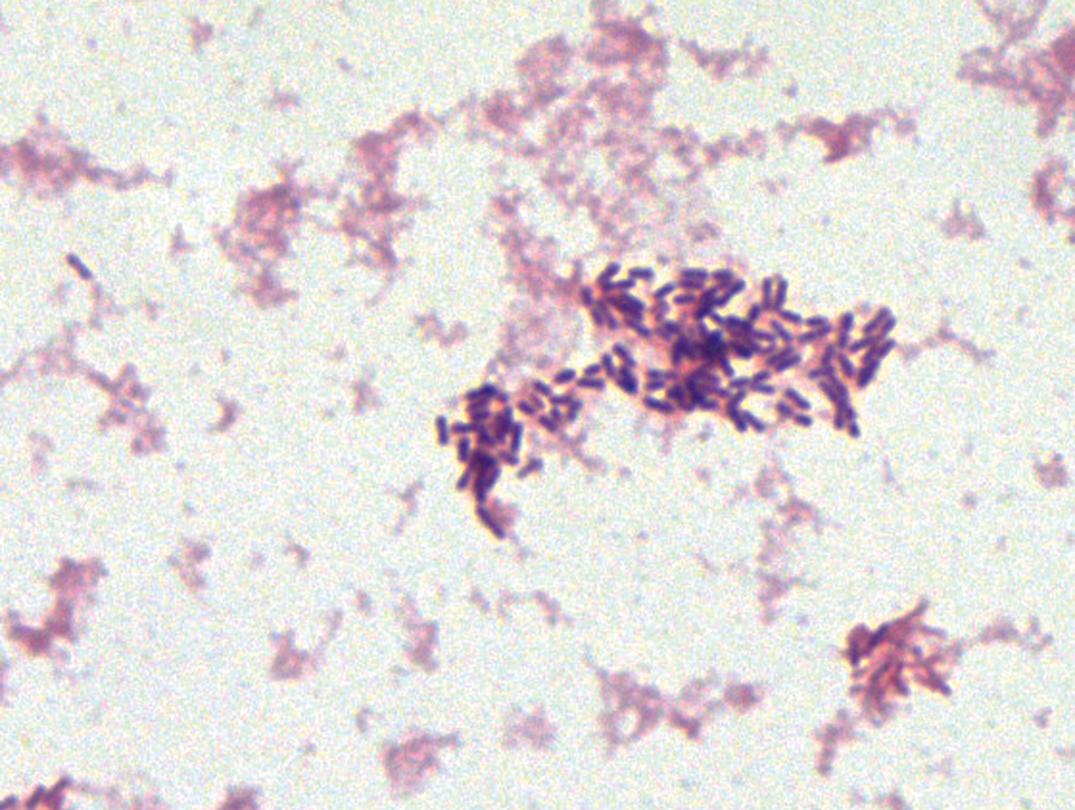
Listeria spp. are nonbranching, non–spore-forming gram-positive rods. L. monocytogenes ( Fig. 57.8 ) is the only species of Listeria that is pathogenic for humans, and L. ivanovii is the only species of the other five Listeria spp. that is pathogenic for animals. Optimal growth of L. monocytogenes is observed between 30°C and 37°C; however, growth may occur as low as 4°C. Colonies are small after 24 hours and exhibit a narrow zone of β-hemolysis on blood agar. A characteristic tumbling motility of saline suspensions of the colonies occurs at room temperature but rarely at 35°C. This same temperature-dependent motility is also noted in semisolid media, in which growth appears as an umbrella shape at the top of the medium (aerobic conditions), preferentially at lower temperatures.
L. monocytogenes is found in soil, dust, water, silage, sewage, and raw unpasteurized milk and in asymptomatic human and animal carriers. Transmission of the organism by foods such as coleslaw, pasteurized milk, soft cheeses, and cantaloupe has resulted in several major epidemics in North America and Europe ( ; ; ; ). According to data from microbiological surveys of food, L. monocytogenes has been detected in 2% to 3% of dairy products, 20% of soft cheeses and processed meats, 30% of certain vegetables (cabbages, radishes), and up to 50% of raw meat and poultry ( ; ). About 1% to 10% of humans are fecal carriers.
Listeriosis is mainly a disease of industrialized countries, occurring sporadically or in epidemics. The primary mode of transmission is contaminated food products, although occasional non–food-related outbreaks have occurred in health care settings, primarily in nurseries, as the result of cross-infection. Contaminated mineral oil for bathing was implicated in one such outbreak ( ). A meta-analysis of over 11,700 literature references pertaining to cases of listeriosis globally resulted in 23,150 illnesses and 5463 deaths in 2010. The proportion of perinatal cases was 20.7% ( ). The incidence rate of listeriosis in the United States was 0.27 cases per 100,000 between 2004 and 2009 and 0.29 per 100,000 from 2009 to 2011; in adults aged over 65 years, the incidence increased to 1.3 cases per 100,000 inhabitants ( ).
Clinical manifestations of listeriosis differ among pregnant women, neonates, and immunocompromised individuals, which constitute the high-risk groups. Listeriosis during pregnancy, most common in the third trimester, presents as a flu-like illness. Bacteremia occurs concomitantly, during which time the uterine contents are infected. Progression to amnionitis may induce premature labor or septic abortion in 3 to 7 days. Infection in the mother is self-limited because the source of infection is removed with delivery of the infected fetus and placenta. Neonatal listeriosis may have an early or late onset. Early-onset disease, manifested at birth or a few days thereafter, results from in utero infection. Infants present with temperature instability, hemodynamic compromise, and respiratory distress. Widely disseminated granulomas, particularly involving the placenta, posterior pharynx, and skin, are characteristic of the illness but are not always present. Late-onset disease, affecting full-term infants of mothers with uncomplicated pregnancies, is assumed to be acquired postpartum, but in most cases the source is unknown. Clinical manifestations of meningitis become apparent several days to weeks after birth.
Nonperinatal listeriosis usually occurs in immunosuppressed individuals, but in about one-third of cases, no risk factor is identified. Tropism for L. monocytogenes for the central nervous system (CNS) is manifested predominantly as meningitis; other forms of CNS listeriosis include cerebritis and brainstem and spinal cord abscesses. Severe disease with high mortality rates (20% to 50%) and neurologic sequelae among survivors are common. Primary bacteremia or focal infections outside the CNS are uncommon. Primary cutaneous listeriosis has occurred occupationally in veterinarians and abattoir workers after exposure to infected animal tissues. Endocarditis, osteomyelitis, arthritis, endophthalmitis, and other focal infections have been reported rarely. Febrile gastrointestinal disease due to L. monocytogenes has been reported in nonimmunocompromised patients; implicated foods include chocolate milk, rice salad, and delicatessen meats and cheeses ( ; ). Immunocompromised patients, such as those with leukemias or bone marrow transplants and patients on immunosuppressive therapies, are cautioned against eating uncooked deli meats to decrease the risk of infection with this organism.
The pathogenesis of listeriosis has been well elaborated in recent years. Host susceptibility, gastric acidity, inoculum size, and virulence properties of the organism, along with specific food products, are the most common factors that determine progression from infection to disease and the severity of that disease in the infected individual. L. monocytogenes can penetrate the epithelial cells of the gastrointestinal tract and grow within hepatic and splenic macrophages. From there, the organism can spread to the CNS or the pregnant uterus. Virulence factors—such as internalin and E-cadherin, a placental receptor—interact to result in infection in the pregnant female/fetus. Immunity to listeriosis relies on T cell–mediated activation of macrophages by lymphokines ( ; ).
Colonies are small and grayish-blue, growing in 24 to 48 hours, and are surrounded by a narrow zone of β-hemolysis on blood agar. A positive catalase reaction differentiates L. monocytogenes from similarly appearing group B streptococci. Organisms are motile at room temperature and produce acid from glucose, trehalose, and salicin, and hydrolyze esculin. Other biochemical characteristics of L. monocytogenes and differences between Listeria and Erysipelothrix are listed in Table 57.3 . Successful use of MALDI-TOF for the identification of L. monocytogenes has been reported ( ).
| Test | L. monocytogenes | E. rhusiopathiae |
|---|---|---|
| β-Hemolysis | + | – |
| Growth at 4°C | + | – |
| Catalase | + | – |
| Motility | + | – |
| Esculin hydrolysis | + | – |
| Gluconate utilization | + | – |
| Voges-Proskauer | + | – |
| H 2 S in triple sugar iron agar | – | + |
L. monocytogenes is usually susceptible to penicillin, ampicillin, erythromycin, chloramphenicol, tetracycline, and gentamicin. Isolates usually are only moderately susceptible to the quinolones. Cephalosporins are ineffective against Listeria spp.; isolates should not be tested against cephalosporins because they are ineffective in vivo regardless of the in vitro result. Methods of performing susceptibility tests for Listeria and interpretive criteria can be found in CLSI document M45-A3 ( ). Resistance to chloramphenicol, macrolides, and tetracyclines has been found in several clinical isolates ( ; ). Ampicillin, alone or in combination with an aminoglycoside, has been used successfully in the treatment of infections caused by L. monocytogenes . Trimethoprim-sulfamethoxazole (TMP-SMX) may be used as alternative therapy in penicillin-allergic patients. Newer gram-positive antibiotics—such as daptomycin, linezolid, and tigecycline—appear susceptible in vitro, but limited clinical data document their in vivo effectiveness.
Erysipelothrix rhusiopathiae is a catalase-negative, non–spore-forming, nonmotile, facultatively anaerobic gram-positive bacillus that has a worldwide distribution. Microscopically, they appear as short rods with rounded ends, occurring singly, in short chains, or in nonbranching filaments. E. rhusiopathiae is a recognized pathogen in humans, occasionally causing erysipeloid, a localized cutaneous infection of hands and fingers obtained after exposure to animals or animal products ( ).
E. rhusiopathiae is usually transmitted to humans from animals by means of skin wounds produced by contaminated objects or in contact with blood, flesh, viscera, or feces of infected animals. E. rhusiopathiae is widespread in nature in wild and domestic animals, birds, fish, and decaying organic matter and causes infection in swine, sheep, rabbits, cattle, birds, and fowl. At risk of infection with this organism are butchers, abattoir workers, fishermen, fish handlers, poultry processors, and veterinarians. The most common form of erysipeloid is a local cutaneous infection manifested by pain, swelling, and a cutaneous eruption characterized by a slowly progressive, slightly elevated, violaceous zone around the site of inoculation. The swelling and erythema migrate peripherally, and the lesion involutes without desquamation. Systemic disease is rare, but numerous case reports describe septicemia and endocarditis. Also rarely reported have been cases of arthritis and brain abscess. Virulence factors include a hyaluronidase, a neuraminidase, and a heat-labile capsule ( ).
Biopsy and tissue aspirates from erysipeloid lesions are the best specimens for culture. The organisms are located deep in the subcutaneous layer of the leading edge of the lesion; therefore, swabs of the surface of the skin are not useful. The organism will grow on blood or chocolate agar, but may require up to 7 days for growth. Conventional blood culture media are suitable for its isolation from blood.
E. rhusiopathiae is oxidase and catalase negative. It is considered nonhemolytic, although greenish discoloration of the media beneath the colonies is often observed after 2 days of incubation. Characteristically, it produces hydrogen sulfide (H 2 S) in triple-sugar iron agar (TSIA) and ferments glucose and lactose. It is nonmotile, does not reduce nitrates to nitrites, and is negative for urease, gelatin, and esculin hydrolysis. A trait highly characteristic of E. rhusiopathiae is the “pipe cleaner” pattern of growth in gelatin stab cultures incubated at 22°C. This organism can be readily distinguished from L. monocytogenes , Lactobacillus , and Bacillus species by MALDI-TOF and biochemical characteristics (see Table 57.3 ) ( ).
Erysipelothrix is susceptible to the penicillins, the cephalosporins, imipenem, erythromycin, clindamycin, chloramphenicol, the tetracyclines, and the fluoroquinolones but is resistant to sulfonamides, aminoglycosides, and vancomycin. Penicillin is the treatment of choice for localized and systemic infection ( ). Methods used for performing susceptibility tests for E. rhusiopathiae and interpretive criteria can be found in CLSI document M45-A3 ( ).
Prevention of human disease is recommended by control of animal disease through sound husbandry, herd management, good sanitation, and immunization procedures ( ).
Members of this genus are strictly aerobic or facultatively anaerobic, rod-shaped, spore-forming, gram-positive, and catalase-positive organisms. Figure 57.9 shows a Gram stain of a Bacillus sp. seen in pleural fluid. With the notable exception of B. anthracis, they are usually motile by means of lateral or peritrichous flagella. They can easily over-decolorize, resulting in gram-variable rods and, because of their variable oxidase reactions, can be confused with gram-negative bacilli. The most reliable diagnostic characteristic of the genus is spore formation, which occurs optimally and on a variety of media under aerobic conditions at 25°C to 30°C. In gram-stained smears, endospores are detectable by the presence of unstained defects or holes within the cell.
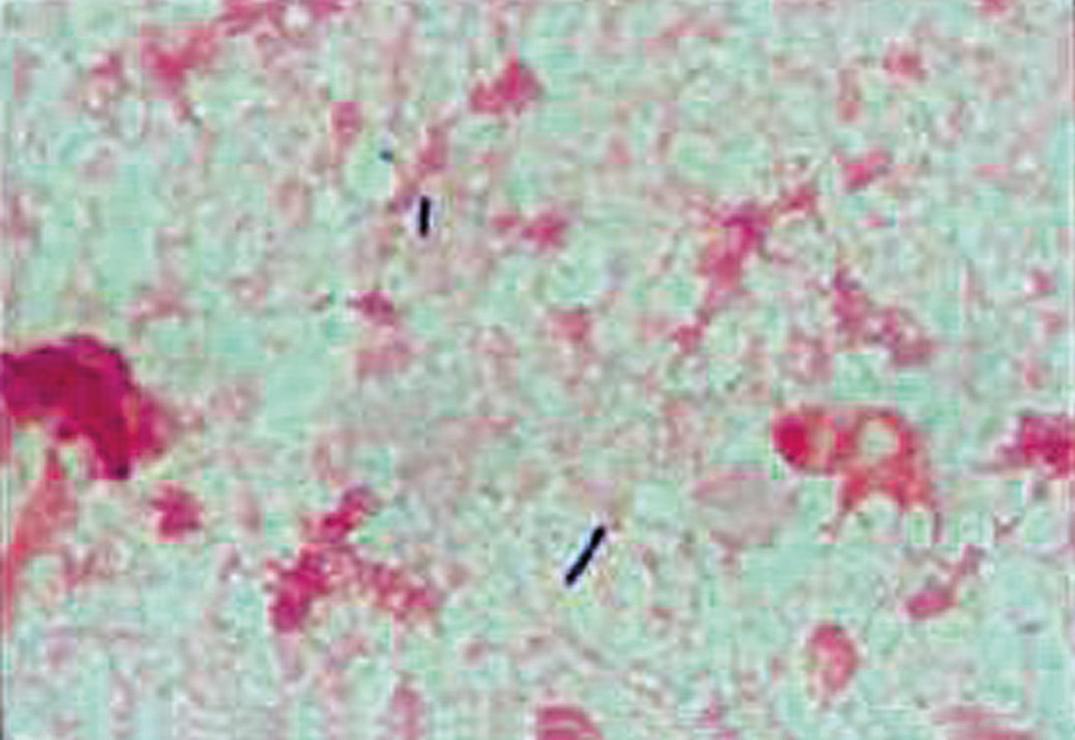
Of the numerous species of Bacillus , B. anthracis is the only one that is uniformly and highly pathogenic. Great care must be exercised when handling material suspected of harboring this species. Work should be performed in biological safety cabinets by gloved, gowned, masked, and immunized personnel; work surfaces must be disinfected with 5% hypochlorite or 5% phenol; and all supplies, materials, and equipment must be decontaminated. Because B. anthracis spores have been used as a means of bioterrorism, cultures containing suspected B. anthracis should be handled only by reference or public health laboratories.
Three forms of anthrax are recognized: cutaneous, inhalation, and intestinal. In its cutaneous form, anthrax produces a small, red, macular lesion that progresses to a vesicle and finally to necrosis, with formation of a characteristic black eschar. Regional lymphadenopathy and septicemia may occur. Mortality in untreated cases with this form of disease is approximately 20%. Inhalation of anthrax spores can lead to acute bronchopneumonia, mediastinitis, and septicemia (“woolsorter’s disease”). The mortality in recognized cases of this form of disease is nearly 100%. Intestinal anthrax follows the ingestion of contaminated food and is manifested by nausea, vomiting, and diarrhea. In some cases, gastrointestinal bleeding is followed by prostration, shock, and death. Septicemia can occur in all three forms of anthrax and may lead to a fatal, purulent meningitis. More recently, a fourth type of anthrax, injectional anthrax, has been recognized in heroin addicts, resulting in severe soft-tissue infections with an increased risk of shock and higher level of mortality than is associated with cutaneous infections ( ).
A major factor in the organism’s pathogenic capabilities is its glutamyl polypeptide capsule, which inhibits phagocytosis; anticapsular antibodies do not protect against the disease. A complex toxin with three components (edema factor, protective antigen, and lethal factor) is responsible for the signs and symptoms of anthrax. The cell wall peptidoglycan is believed to produce a robust intravascular inflammatory response that can lead to hemodynamic and organ dysfunction, resulting in shock ( ; ).
Humans become infected with anthrax by contact with and inhalation or ingestion of infected animals, their carcasses, or their by-products. Cattle, sheep, horses, and goats are the animals most frequently infected and provide a ready source of vegetative organisms that sporulate and perpetuate the environmental contamination.
Although usually saprophytic, other species of Bacillus can cause disease. B. cereus has been associated with ear infection, pneumonia, posttraumatic ocular wound infection, septicemia, endocarditis, and meningitis. Patients with pneumonia and septicemia are often immunosuppressed. Bacteremia is frequently associated with intravenous drug use and with contaminated intravascular devices ( ).
Two forms of gastroenteritis are associated with Bacillus species. Food poisoning caused by Bacillus may occur within 1 to 6 hours following ingestion of food contaminated by B. cereus that has produced a preformed heat-stable toxin. Major manifestations of Bacillus food poisoning include nausea, vomiting, cramps, and occasionally diarrhea, but no fever. Typically, this form of Bacillus gastroenteritis results from the bulk preparation of foods that are not reheated before they are served. B. cereus type 1 grows particularly well in fried rice and is more heat resistant than other types; thus, this form of gastroenteritis is frequently seen in association with consumption of cooked rice in Chinese restaurants. The second form of gastroenteritis caused by Bacillus spp. results from contamination of poultry and vegetable dishes and is characterized by the onset of cramps and diarrhea 8 to 16 hours following ingestion of contaminated food. In this instance, the major manifestations of B. cereus infection are caused by the production of a heat-labile enterotoxin. From 1998 to 2008, 1229 food-borne outbreaks caused by B. cereus (rice dishes were implicated in 50% of the cases), Clostridium perfringens , and S. aureus were reported in the United States. In 2008, there were 17 to 18 suspected outbreaks of gastroenteritis associated with B. cereus as compared to over 40 outbreaks due to S. aureus and 10 or 11 outbreaks due to C. perfringens ( ).
The genus Bacillus contains more than 300 species; other than B. anthracis and B. cereus , common species include B. subtilis , B. licheniformis , B. megaterium, B. pumilus , and B. thuringiensis . Many species have been renamed, however, and more than 100 new genera of gram-positive spore-producing aerobic bacilli have been named. One of those genera, Paenibacillus, contains species that have been associated with clinical disease, including meningitis and endophthalmitis. Species of Paenibacillus have been implicated in infections associated with penetrating injuries and contaminated medical devices ( ; ; ). P. macerans has been found in contaminated blood culture bottles in a neonatal intensive care unit, and P. pasadenensis has been isolated in spacecraft facilities and also as a cause of mediastinitis following heart surgery ( ; ).
In cases of suspected cutaneous anthrax, vesicle fluid and material under the edge of the eschar should be collected with a swab for smear and culture. For suspected inhalation anthrax, sputum should be collected for smear and culture. Oropharyngeal and rectal swab cultures should be considered in the intestinal form. Smears and cultures of CSF should be requested in suspected meningitis. In the septicemic stage, blood cultures should be performed. Finding large, boxcar-shaped gram-positive cells in smears of any of these specimens should raise suspicion for the diagnosis.
Species of Bacillus grow well on sheep blood agar. Colonies of B. anthracis are usually flat, with an irregular margin (“Medusa head”), appear off-white with a ground-glass surface, and are usually nonhemolytic. Figure 57.10 demonstrates the colonial morphology of B. anthracis on blood agar. When touched with an inoculating loop, the colonies are tenacious and will stand up like a beaten egg white. The Medusa head colony may be seen with B. cereus and certain other Bacillus species. Anthrax bacilli are nonmotile in a hanging drop test or in semisolid media, whereas most other species of Bacillus are motile. Motility by the hanging drop method is a useful differential test between B. anthracis and B. cereus but must be carried out with a fresh broth culture of the organism. As mentioned earlier, because of the use of B. anthracis in terrorist attacks, sentinel laboratories (level A) should send any suspicious isolates of this organism to their public health laboratory or the CDC. A commercial system has been evaluated recently for identification of aerobic endospore-formers; 93% of the strains were correctly identified to species level in the genera Bacillus , Paenibacillus , Aneurinibacillus , Brevibacillus , Geobacillus , and Virgibacillus ( ). Bacillus spp. other than anthracis have been correctly identified using MALDI-TOF ( ).
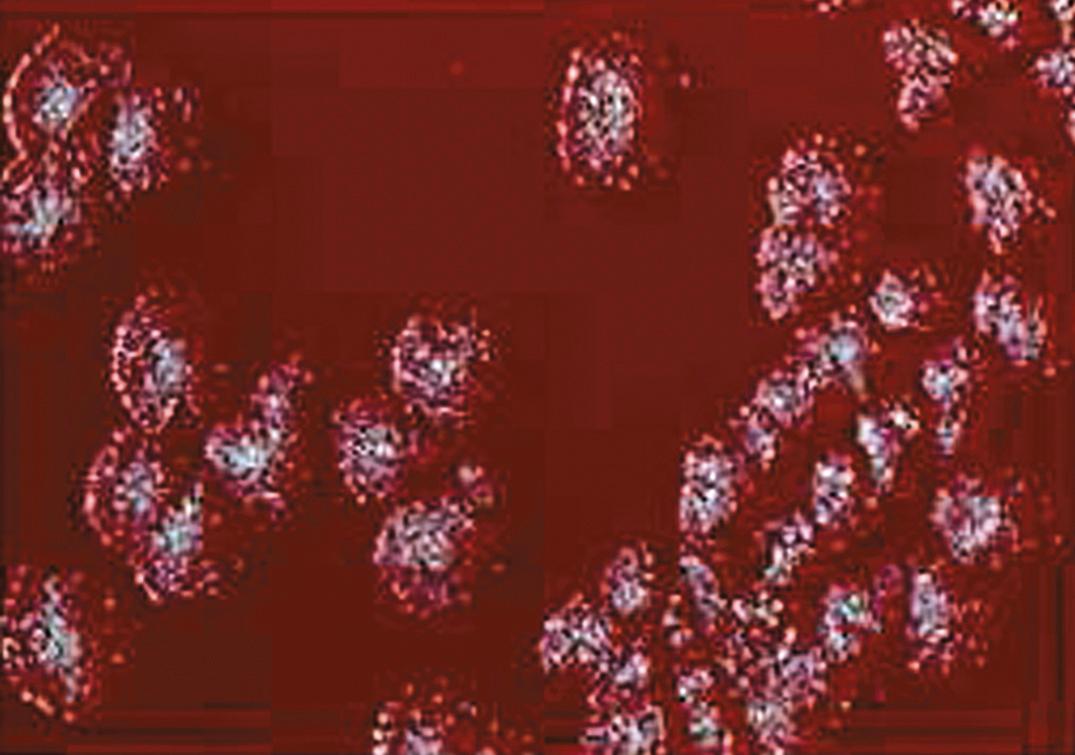
The diagnosis of B. cereus gastroenteritis cannot be accurately made by culture of stool because the organisms may be a component of the indigenous gut flora. Diagnosis, therefore, depends on quantitative culture of the suspected contaminated food. The presence of greater than or equal to 10 5 CFUs/g in the suspected food constitutes presumptive evidence of B. cereus food poisoning (Logan, 2011). This testing is usually not performed in a routine clinical microbiology laboratory.
Although susceptible to a variety of agents, antibiotic therapy of anthrax has centered on the use of fluoroquinolones. These agents are highly active against B. anthracis , strains of which are typically susceptible to β-lactam antibiotics. However, many strains of Bacillus spp. elaborate β-lactamases in nature; thus, these agents usually are not considered as first-line drugs of choice until the specific susceptibility of the isolate is known. Most strains of Bacillus spp. are inhibited by fluoroquinolones, tetracyclines, aminoglycosides, and chloramphenicol at low concentrations ( ) and most are susceptible to vancomycin, but reports have described vancomycin resistance in B. circulans and P. thiaminolyticus. Bacillus strains have been demonstrated to carry genes similar to the vanA gene of the enterococci ( ). Methods of performing susceptibility tests and of interpreting their results can be found in the third edition of the CLSI document M45 ( ).
Prevention of anthrax in humans ideally depends on its control in animals. Prompt diagnosis of sick animals, their isolation and therapy, and cremation of carcasses are indicated when sporadic outbreaks occur. In enzootic areas, vaccination with nonencapsulated spore preparations is used. Occupationally exposed persons should also be immunized. Acute diarrheal disease caused by B. cereus may be prevented by proper cooking and refrigeration of foods prepared in bulk to prevent proliferation of vegetative forms of the bacteria and formation of the enterotoxin.
In gram-stained smears of clinical specimens, Nocardia spp. appear as long, thin, beaded, branching gram-positive bacilli ( Fig. 57.11 ). The most distinguishing quality of the nocardiae is their partial acid fastness; cells stain positively with a modified acid-fast (Ziehl-Neelsen or Kinyoun) stain, differentiating them from Actinomyces , which may have a similar gram-stain appearance. Partial acid fastness may be difficult to demonstrate and can be enhanced by growing the organism for about 4 days on Middlebrook 7H10 agar or in litmus milk broth. Most clinical infections have been caused by N. abscessus , N. brasiliensis , N. cyriageorgica , N. farcinica, N. nova , N. pseudobrasiliensis, N. veterana , and N. wallacei ( ; ).
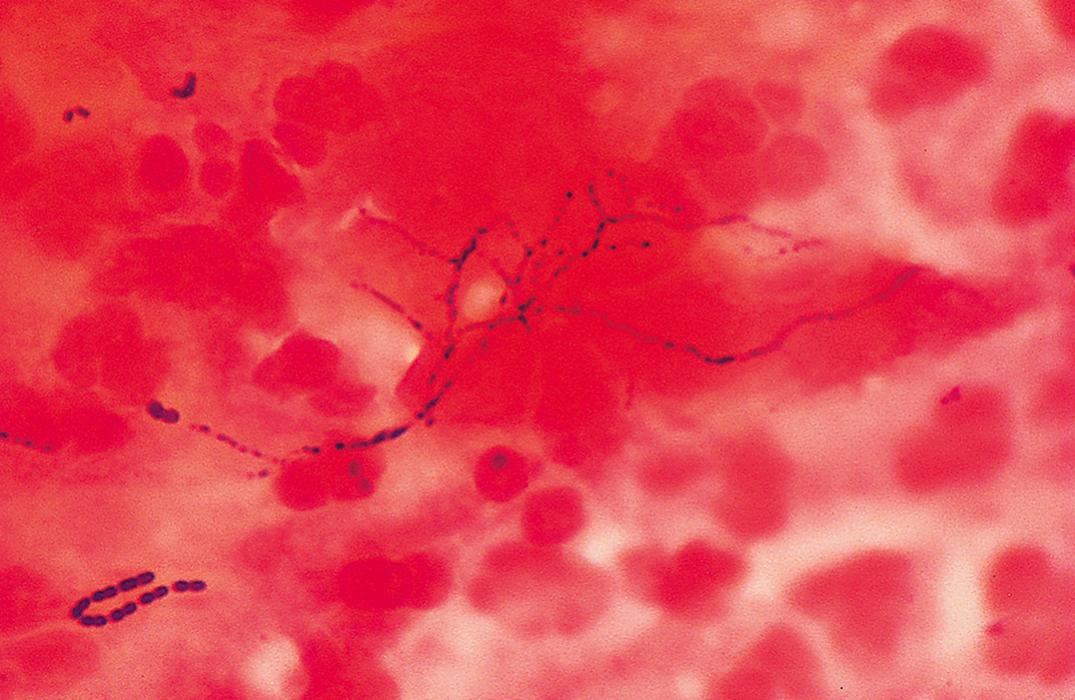
Nocardiae are found in soil and organic material worldwide, causing disease in many animals and in fish. It is usually acquired via inhalation of the organism but may occur following trauma and contact with contaminated soil, or the organism may enter the body via the gastrointestinal tract when contaminated material contacts an area of mucosal ulceration.
In the lungs, Nocardia spp. are phagocytosed by alveolar macrophages and grow intracellularly, eliciting a mixed inflammatory response (neutrophils, lymphocytes, and macrophages), eventually resulting in abscess or, occasionally, granuloma formation. In vitro studies of the host defenses against Nocardia spp. suggest that neutrophils, activated macrophages, and cytotoxic T cells are involved. Although neutrophils do not kill virulent Nocardia spp., they inhibit their growth, possibly suppressing the infection until macrophages are fully activated. If the infection is not contained within the lung, organisms spread to other tissues by advancing growth, thus producing empyema, chest wall involvement, and draining sinuses, or by hematogenous dissemination, resulting in abscess formation, especially in the brain, subcutaneous tissues, and kidneys. The primary host factor associated with increased risk of nocardiosis is cellular immune dysfunction, although many persons infected with Nocardia spp. have no recognized cellular or humoral immune defect ( ; ).
Pulmonary disease, the most frequent manifestation of nocardiosis, is characterized by fever, anorexia, weight loss, cough, dyspnea, and pleuritic chest pain. Skin and subcutaneous disease may present as pyoderma, cellulitis, single or multiple abscesses, lymphocutaneous disease resembling sporotrichosis, or nodules. In Central and South America, primary infections of the skin with N. brasiliensis may produce an actinomycetoma (a localized indurated granulomatous mass with sinus tracts draining pus and “sulfur” granules), typically on the lower extremities. Disseminated disease is usually caused by members of the N. asteroides complex. It originates in the lung in most cases and typically is manifested as single or multiple abscesses involving the CNS ( ). The skin is the second most common site of dissemination, followed by kidney, liver, and lymph nodes (Conville & Witebsky, 2011).
Nocardia spp. grow aerobically on most nonselective media, such as sheep blood and chocolate agars, potato dextrose agar, Sabouraud dextrose agar, and Löwenstein-Jensen or Middlebrook media and in 7H9 broth used for mycobacterial culture. These organisms will grow on buffered charcoal-yeast extract (BCYE) agar used for the isolation of Legionella spp. as well. It is important to note that Nocardia spp. may not always survive the decontamination procedures used for recovery of mycobacteria. Incubation in the presence of 10% CO 2 enhances growth. Nocardia spp. may grow in 48 hours, but colonies typically appear in 5 to 10 days as waxy, bumpy, or velvety rugose forms, often with yellow to orange pigment, depending on the species. Observing branching filaments that stain only with a modified acid-fast stain distinguishes Nocardia spp. from mycobacteria. Nocardia spp. are differentiated from most other aerobic actinomycetes by testing resistant to the action of lysozyme ( Nocardia spp. are resistant; Streptomyces spp., Rhodococcus spp., Gordona spp., and Actinomadura spp. are susceptible) and examining their morphology on tap water agar. For example, the latter can help in differentiating the branching Nocardia from nonbranching Rhodococcus spp. Because many new species of Nocardia have been recognized over the past several years, the use of biochemical tests (e.g., casein, hypoxanthine, tyrosine, xanthine) is not sufficient for identification. Molecular tests (e.g., 16S rDNA sequencing, PCR-restriction enzyme pattern analysis [PRA]) of MALDI-TOF ( ) are required for accurate species identification ( ; ). Identification to the species level is important because of the variability in antibiotic susceptibility patterns noted among species ( ; ).
Sulfonamides (alone or in combination with trimethoprim—e.g., TMP-SMX) are usually the antimicrobial agents of choice. However, optimal antimicrobial therapy depends on the species of Nocardia present, the susceptibility pattern of the individual strain, and the type of infection. Other potential antimicrobial agents include amikacin, clarithromycin, imipenem, or a quinolone, depending on the species isolated. The CLSI has information about methods used for susceptibility testing and interpretation of results for Nocardia and other aerobic actinomycetes ( ).
Other genera of actinomycetes that are medically relevant to humans include Rhodococcus , Gordonia , Tsukamurella , Actinomadura , and Streptomyces . One other member, Tropheryma whippelei , is nonculturable and is the causative agent of Whipple disease.
Members of this group of organisms are ubiquitous in the environment and have been isolated from soil, fresh water, marine water, and organic matter. R. equi is the most common Rhodococcus sp. pathogenic to humans. It is an opportunistic pathogen in severely immunocompromised hosts, causing a slowly progressive granulomatous pneumonia. It may be isolated from the blood of infected patients. The organism is likely acquired via the respiratory route, possibly as a result of exposure to infected animals. The organism’s ability to persist in and ultimately destroy macrophages probably accounts for its ability to cause disease ( ). Infections caused by Gordonia spp. and Tsukamurella spp. are increasingly being reported ( ; ). Tsukamurella spp. appear to be pathogenic only when certain clinical conditions are present (e.g., immunosuppression, presence of foreign body, active chronic infection, such as tuberculosis) ( ). Actinomadura spp. are a frequent cause of actinomycotic mycetomas, a chronic infection of skin and subcutaneous soft tissues. Most of these infections are seen in tropical and subtropical countries, where walking barefoot increases the chance of exposure through repeated puncture wounds. Streptomyces spp. have traditionally been considered of little medical significance; however, one species, S. somaliensis , has been identified as an etiologic agent of actinomycotic mycetoma. Other Streptomyces spp. have only occasionally been reported to be of medical importance ( ).
Aerobic actinomycetes are slow growing and may require 2 to 3 weeks of incubation. These microorganisms grow on most of the nonselective media used to isolate bacteria, mycobacteria, and fungi. Species of Rhodococcus grow as coccobacilli in a zigzag fashion. Rudimentary branched filaments have been observed from liquid media. Colonies may be rough, smooth, or mucoid and have pigments ranging from buff to coral to orange to deep rose after several days of incubation. R. equi is usually pale pink, pale yellow, or coral and may appear slimy. Gordonia spp. range from smooth, mucoid colonies that are adherent to the media to dry, raised colonies. The pigment produced may be beige to salmon colored. Tsukamurella spp. are slightly acid fast by the modified Kinyoun. They appear as long rods that fragment into three parts. No aerial hyphae are produced. The colonies are circular, with entire or rhizoid edges. They may be dry or creamy, with a white to orange pigment. Rough colonies may be produced after 7 days of incubation. Species of Actinomadura form waxy, cerebriform, tough, membranous white, yellow, pink, or red colonies. Colonies of Streptomyces are dry to chalky, heaped or folded, gray-white to yellow, and have the odor of a musty basement. A wide variety of pigments are produced that color the substrate and hyphae. Some species do not produce aerial hyphae. As with the Nocardia spp., complete identification of these members of the aerobic actinomycetes often requires molecular sequencing methods or MALDI-TOF ( ; ).
This genus is composed of species that are nonmotile and catalase and oxidase positive. These aerobic, gram-negative cocci are often arranged in pairs, with flattened adjacent surfaces, giving the appearance of kidney beans or coffee beans ( Fig. 57.12 ). The organisms are somewhat fastidious, in some instances requiring the addition of blood, serum, cholesterol, or oleic acid to the medium to counteract growth inhibitors, such as fatty acids. N. gonorrhoeae and N. meningitidis generally require prompt incubation in CO 2 for growth. However, this is strain dependent, varies with the phase of the organism’s growth curve, and is often lost in subcultures. N. meningitidis and most N. gonorrhoeae are not inhibited by the presence of vancomycin or lincomycin, colistin, and nystatin—a characteristic that is particularly useful in their selective isolation from specimens contaminated by other bacteria ( ).
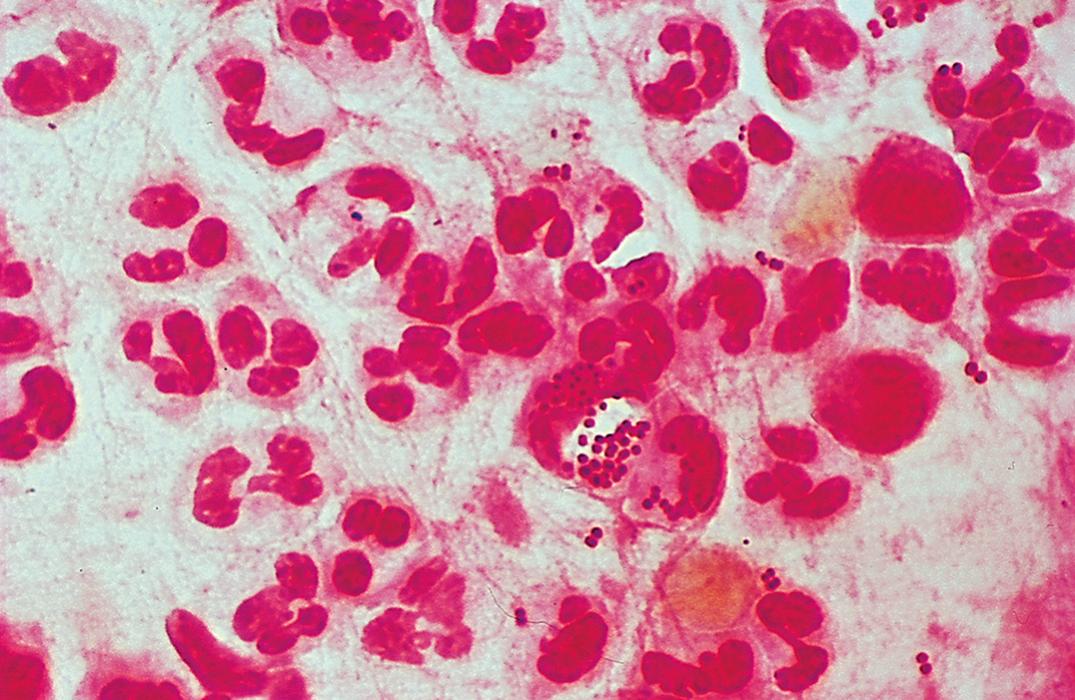
Although opportunistic infections caused by species of Neisseria other than N. gonorrhoeae and N. meningitidis have occasionally been reported in compromised hosts, these species are generally nonpathogenic. N. meningitidis may colonize the mucous membranes of the upper respiratory tract—an event that is usually followed in 7 to 10 days by the formation of bactericidal and hemagglutinating antibodies, which may not eliminate the carrier stage but convey group-specific immunity. In a few cases, disease results shortly after colonization, most frequently in the form of meningococcemia and meningitis. The organism also has a tendency to invade serous membranes and joint tissues, with the development of pleuritis, pericarditis, and arthritis. Carriage of N. meningitidis in the nasopharynx occurs in 5% to 15% of healthy individuals; this rate may be higher in confined groups such as military recruits. A direct correlation between carrier rates and incidence of disease has not been established, with the possible exception of members of large households or households with an infant or childhood case during epidemics of disease. N. meningitidis has also been isolated from genital sources, where its clinical significance is uncertain. When cultured from these sources, bacteria may be misidentified as N. gonorrhoeae unless appropriate tests for distinguishing these species are performed.
N. gonorrhoeae is always considered a pathogen, even in the absence of obvious clinical symptoms. In the United States, an estimated 820,000 new N. gonorrhoeae infections occur each year, and gonorrhea is the second most commonly reported communicable disease ( ). Urethral infections caused by N. gonorrhoeae among men produce symptoms that cause them to seek curative treatment soon enough to prevent sequelae but often not soon enough to prevent transmission. In male patients, local complications of urethral gonorrhea include epididymitis, penile edema, and abscesses of the urethral and/or bulbo-urethral glands.
In females, N. gonorrhoeae causes infection of the endocervix. Infections of the urethra, rectum, and the Bartholin and Skene glands can also occur. Asymptomatic infections due to N. gonorrhoeae are more common in female patients. Typical symptoms of N. gonorrhoeae infection in women include increased vaginal discharge, dysuria, and intermenstrual bleeding. Complications of untreated infection include ascension of the infection, potentially resulting in pelvic inflammatory disease (PID), including salpingitis, tubo-ovarian abscesses, endometritis, and peritonitis. PID can result in tubal scarring that can lead to infertility or ectopic pregnancy. Up to 80% of infected women are asymptomatic and, therefore, untreated. The lack of therapy may allow dissemination of the infection, manifesting as skin lesions and/or arthritis. Disseminated gonococcal infection (DGI) is relatively uncommon (0.5%–3.0%) but should be considered when infective arthritis occurs in young people. Two clinical presentations of DGI are typical: an early-stage polyarthritis/dermatitis and a later-stage septic arthritis. The dermatologic manifestations include hemorrhagic pustules and papules; hemorrhagic bullae or necrotic lesions can be seen. Polyarthritis of the knees and elbows is typical.
The principal virulence factor of N. meningitidis is a lipopolysaccharide–endotoxin complex. In experimental animals, it activates the clotting cascade, depositing fibrin in small vessels, producing hemorrhage in the adrenals and other organs, altering peripheral vascular resistance, which leads to shock and death. This can initially present as petechial lesions, which can coalesce and become ecchymotic. The pathogenesis and clinical manifestations of N. gonorrhoeae infections differ somewhat from those of N. meningitidis . Pathogenic types of N. gonorrhoeae adhere by means of pili, which are not produced by nonpathogenic types, to various human cells. These antigenically heterogeneous pili, which represent one of the principal virulence factors of N. gonorrhoeae , may inhibit phagocytosis and stimulate strain-specific antibody formation. Other possible virulence factors of N. gonorrhoeae are less clearly defined. Both N. gonorrhoeae and N. meningitidis produce an IgA protease, which may be important in their pathogenesis because IgA is the antibody class that predominates in secretions on mucous membranes ( ).
The single most important element in the laboratory diagnosis of infection caused by N. meningitidis or N. gonorrhoeae is the specimen, including proper selection, collection, and transport to the laboratory. The pathogenic species are sensitive to drying and extremes of temperature, and material must be cultured promptly to enhance recovery. They are mesophilic and grow poorly, if at all, at room temperature. Many require prompt incubation in CO 2 (2%–8%) for primary isolation.
Chocolate media are commonly used for cultures of N. gonorrhoeae and should contain antibiotics (i.e., vancomycin or lincomycin, as well as colistin, nystatin or anisomycin, and trimethoprim) if the specimen is prone to be contaminated by normal flora. Vancomycin-susceptible gonococci will grow on media containing lincomycin. However, because of the synergistic interaction of lincomycin and trimethoprim, the latter must be omitted from media containing lincomycin. Direct inoculation of specimens at the bedside followed by prompt incubation at 35°C in CO 2 is optimal. This can be accomplished in several ways: placing the medium into a candle jar; placing the medium in a sealed bag containing a citric acid bicarbonate tablet; or using a medium contained within a bottle having a CO 2 atmosphere. If any of these culture systems must be mailed to a reference laboratory for processing, they must first be incubated overnight to ensure growth of the organisms.
An isolate from a urogenital specimen showing the appropriate colony appearance on a selective medium presumptively may be called N. gonorrhoeae based on results of Gram stain and oxidase and catalase tests. Gram-stained smears prepared from colonies of N. gonorrhoeae should show typical gram-negative diplococci, but organisms may occur in tetrads, especially from young cultures. All species of Neisseria are oxidase positive, and all species except N. elongata are catalase positive. Because Neisseria other than N. gonorrhoeae may be recovered from urogenital sites, confirmatory testing is strongly recommended and is required for all isolates from extragenital sites and when sexual abuse is suspected (preferably by more than one method).
Confirmation of N. gonorrhoeae and identification of the other Neisseria spp. are based on growth and biochemical characteristics ( Table 57.4 ) ( ). The standard method of identification consists of detection of acid production from carbohydrates in a cystine trypticase acid (CTA) base medium and other conventional biochemical tests. However, given the drawbacks of conventional methods, more rapid identification tests are used in most clinical laboratories. Tests for direct detection of N. gonorrhoeae and N. meningitidis in clinical specimens are also available. Typing of isolates of N. gonorrhoeae and N. meningitidis is done primarily for epidemiologic studies.
| N. gonorrhoeae | N. meningitidis | N. cinerea | N. lactamica | M. catarrhalis | |
|---|---|---|---|---|---|
| Growth | |||||
| Thayer-Martin medium | + ∗ | + | – | + | † |
| Nutrient agar, 25°C | – | – | – | – | + |
| Oxidase | + | + | + | + | + |
| β-Galactosidase | – | – | – | + | – |
| Reduction of nitrate | – | – | – | – | + |
| DNase | – | – | – | – | + |
| Production of Acid from | |||||
| Glucose | + | + | – ‡ | + | – |
| Maltose | – | + | – | + | – |
| Lactose | – | – | – | + | – |
| Sucrose | – | – | – | – | – |
| Fructose | – | – | – | – | – |
∗ Most vancomycin-susceptible strains will not grow on Thayer-Martin medium.
† Some strains positive and others negative.
‡ Weak reaction may occur in rapid carbohydrate utilization tests.
With the standard method of identification, acid production from glucose, maltose, lactose, sucrose, and fructose in a CTA base medium and a carbohydrate-free control are tested. Tubes are inoculated, incubated at 35°C to 37°C in ambient air, and examined at 24-hour intervals until reactions are interpretable, or for 72 hours. Expected results for the species of Neisseria are shown in Table 57.4 . Occasionally, however, an isolate of N. meningitidis yields aberrant carbohydrate reactions: glucose-negative, maltose-negative, or asaccharolytic. If N. meningitidis is strongly suspected in these cases, identification can be confirmed by slide agglutination using pooled polyvalent grouping antisera or sera specific for individual serogroups. In addition to conventional carbohydrate degradation tests, reduction of nitrates and nitrites and production of DNase should be evaluated. The latter is especially useful for identification of Moraxella catarrhalis , which is DNase positive ( Neisseria spp. are DNase negative). Drawbacks to conventional tests are the requirement for a heavy inoculum, the need to work with pure cultures, long turnaround time, and failure of some fastidious strains of N. gonorrhoeae to grow.
Several commercial systems detect acid production from carbohydrates, usually in 1 to 4 hours ( ). The inoculum must be prepared from a pure culture of the isolate; thus, identification is generally available 24 hours after isolation. Acid reactions of some N. gonorrhoeae and, to a lesser extent, N. meningitidis may be difficult to interpret or may be aberrant with some kits, and strains of N. gonorrhoeae that are weak producers of acid from glucose may appear to be glucose negative. Some strains of N. cinerea, which does not typically produce acid from glucose, can appear glucose positive in certain systems.
Enzyme substrate tests provide rapid identification (1–4 hours) only of isolates of oxidase-positive, gram-negative diplococci recovered on a selective medium ( ). They are valuable for differentiating maltose-negative strains of N. meningitidis from N. gonorrhoeae . However, color changes may be subtle and if misinterpreted could cause isolates of N. meningitidis and other Neisseria spp. to be incorrectly called N. gonorrhoeae . Moreover, strains of N. cinerea and Kingella denitrificans that grow on gonococcus-selective media could be misidentified as N. gonorrhoeae if not confirmed by other procedures. Commercial products that combine enzyme substrate tests with modified conventional tests provide accurate identification of species of Neisseria and Haemophilus . Immunologic tests for N. gonorrhoeae —in particular, coagglutination—can be used to confirm the biochemical identification of N. gonorrhoeae . Both false-positive and false-negative results have been reported with these reagents. MALDI-TOF is able to identify both N. meningitidis and N. gonorrhoeae .
Culture detection of N. gonorrhoeae from urogenital sites has been largely replaced by NAAT. Many NAATs are available that simultaneously detect N. gonorrhoeae and Chlamydia trachomatis in vaginal, cervical, urethral, and urine samples with greater sensitivity than culture, and for which loss of viability of N. gonorrhoeae in particular is an issue. These tests are recommended by the CDC as the preferred test in symptomatic and asymptomatic individuals, although there are also recommendations that culture be available for both N. gonorrhoeae and C. trachomatis at least in some laboratories in situations of potential antimicrobial resistance and in some sexual assault situations ( ). These molecular techniques are described in other sections of this text.
Despite the occasional recovery of N. meningitidis strains with decreased susceptibility to penicillin, penicillin G remains the drug of choice for treatment of meningococcal meningitis ( ). Decreased susceptibility of N. meningitidis to penicillin is thought to be due to decreased binding of penicillin by altered meningococcal cell wall penicillin-binding proteins PBP-2 and PBP-3. Other species of Neisseria have also demonstrated this lowered affinity to penicillin. Other agents that have good activity against N. meningitidis include the extended-spectrum cephalosporins and chloramphenicol. Rifampin and ciprofloxacin may be used for prophylaxis among household contacts. Standardized methods of susceptibility testing and breakpoints in the CLSI documents are available ( ). The CLSI recommends broth microdilution or agar dilution with cation-supplemented Mueller-Hinton Broth (with 2%–5% laked horse blood) or Mueller-Hinton agar (with 5% [vol/vol] sheep blood) if testing is done. In laboratories where susceptibility testing for N. meningitidis is not available, β-lactamase testing can be performed by using the chromogenic cephalosporin test, the cefinase nitrocefin disk test, if lowered susceptibility or clinical failure on penicillin is suspected. If positive, isolates can be shipped to a reference laboratory for further testing.
Because of widespread resistance of N. gonorrhoeae to penicillin and tetracycline, the current recommendation for treatment is ceftriaxone plus azithromycin. Although no resistance to the cephalosporins is apparent, resistance to the fluoroquinolones has been documented ( ). Therefore, susceptibility testing should be performed if symptoms persist after treatment. The CLSI document recommends the agar dilution method or a disk diffusion method for testing of N. gonorrhoeae ( ). For some agents, only a susceptible breakpoint is available because no resistant strains have yet been documented. If an isolate with a nonsusceptible result to a third-generation cephalosporin is identified, for example, confirmation tests and referral to a reference laboratory should be strongly considered ( ).
Two meningococcal conjugate vaccines against N. meningitidis serogroups A, C, Y, and W135 are licensed in the United States and recommended for all adolescents at 11 to 12 years and 16 years ( ). There are also recommendations for some adult populations, such as microbiologists who routinely work with N. meningitidis ( ). There is also a conjugate vaccine against serogroup B, which is available for all adolescents and young adults, but specifically recommended for asplenic patients and patients with a complement deficiency. Antibiotic prophylaxis should be limited to household contacts and those who have had contact with patients’ oral secretions. Rifampin is currently the drug of choice. Laboratory safety is very important when handling specimens and cultures positive for N. meningitidis . wrote an article on the appropriate precautions for handling specimens and steps to take if accidents do occur.
The use of preexposure antibiotics to prevent gonococcal disease is discouraged because of the potential risks of sensitization and the emergence of resistant strains. The sole exception to this rule is the application of erythromycin ointment to the eyes of newborns to prevent gonococcal (and chlamydial) ophthalmia ( ).
M. catarrhalis may be carried in the oropharynx of healthy children and adults. It is an encapsulated organism, and extending from its outer membrane are pili that serve as adhesins. The most common infections caused by this organism are bronchitis, otitis, sinusitis, and pneumonia (especially in persons with underlying chronic lung disease) ( ; ). More recently, molecular mechanisms have been described that demonstrate the virulence factors possessed by strains of M. catarrhalis responsible for otitis media ( ). M. catarrhalis is an infrequent cause of bacteremia, endocarditis, meningitis, urogenital infection, and ophthalmia neonatorum.
Figure 57.13 shows the Gram stain of sputum in which M. catarrhalis is seen in large quantities inside and outside the polymorphonuclear leukocytes. M. catarrhalis initially was part of the Neisseria genus and later was transferred to the Branhamella genus for a short time. M. catarrhalis is a coccus that morphologically resembles Neisseria spp., unlike other members of the genus Moraxella (e.g., M. lacunata , M. osloensis, M. atlantae ), which appear as rods. M. catarrhalis bacteria are oxidase and catalase positive but can be differentiated from Neisseria spp. in their ability to grow readily on blood agar, their lack of oxidative metabolism (sugars will be negative), and their production of DNase. Nearly all isolates of M. catarrhalis produce β-lactamase, which can be detected using nitrocefin. Although they should be assumed to be resistant to penicillin because of this, these isolates generally remain susceptible to cephalosporins, TMP-SMX, and β-lactamase inhibitor combinations ( ).
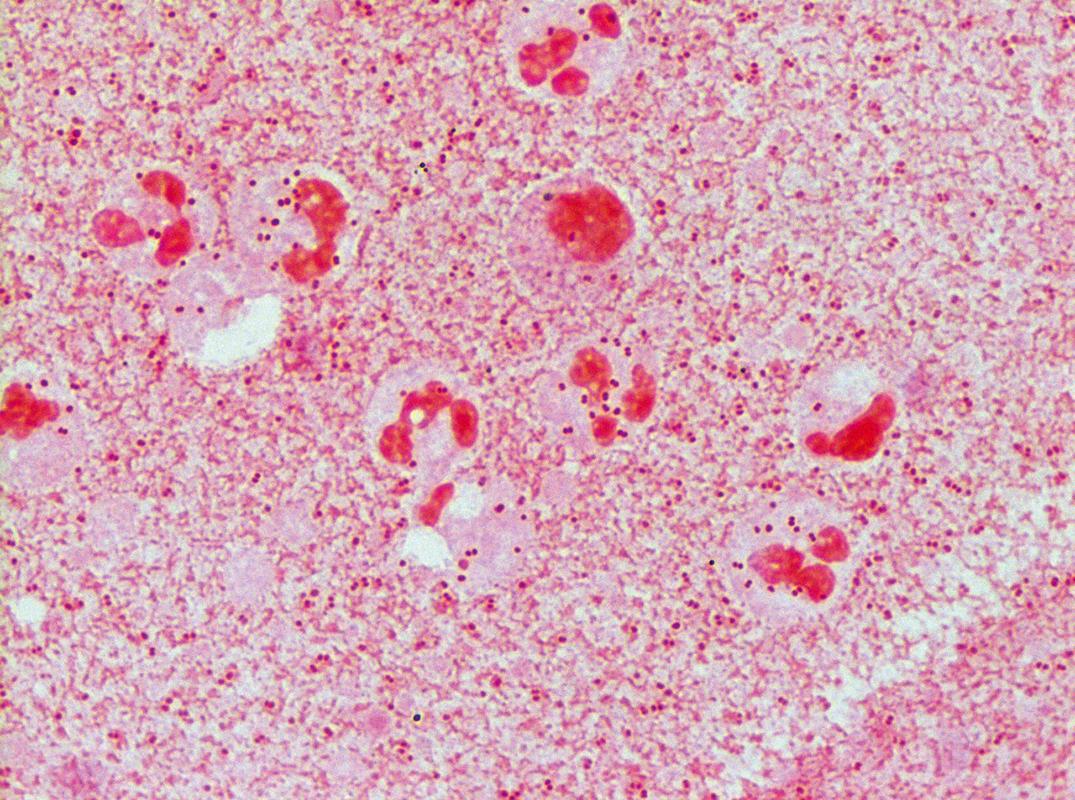
The gram-negative bacilli make up a complex group. They are broken down into Enterobacterales, which are those found normally in the gastrointestinal (GI) tract or that colonize and cause infection there primarily; nonfermentative gram-negative bacilli, which usually are not found as part of the normal flora of humans but rather are environmental bacteria; non-Enterobacterales, which can cause GI infection, such as Vibrio or Campylobacter ; agents of infections with specific epidemiologic characteristics, such as Legionella and Francisella ; other gram-negative bacilli, including Haemophilus spp.; and miscellaneous genera. A good overview of how to approach the identification of the aerobic gram-negative rods can be found in .
The Enterobacterales are facultatively anaerobic, non–spore-forming, nonmotile or peritrichously flagellated, oxidase-negative, gram-negative bacilli that produce acid fermentatively from glucose and reduce nitrates to nitrites. Figure 57.14 shows a Gram stain of Escherichia coli , but it could represent any member of the Enterobacterales. Common clinical genera included in this group are Citrobacter , Enterobacter , Escherichia , Klebsiella , Morganella , Proteus , Providencia, Salmonella , Serratia , Shigella , and Yersinia ( ).
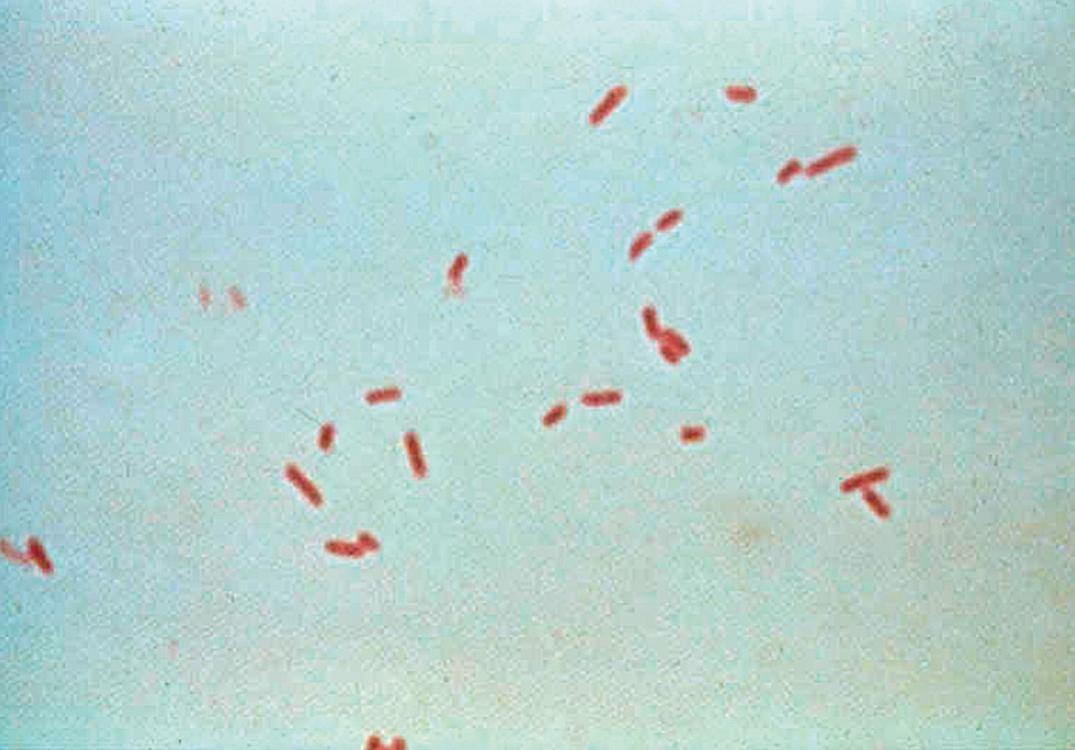
Enterobacterales are found on plants, in soil, in water, and in the intestines of humans and animals. They have been associated with many clinical infections, including abscesses, pneumonia, meningitis, septicemia, and urinary tract infections. Those commonly associated with human infection include E. coli , Klebsiella spp., Proteus spp., Enterobacter spp., Salmonella spp., Shigella spp., Serratia spp., Citrobacter spp., and Providencia spp. In the urinary tract, those frequently isolated are E. coli , Proteus mirabilis , Klebsiella pneumoniae , and K. oxytoca . Gram-negative pneumonias associated with the Enterobacterales are frequently caused by K. pneumoniae . Gram-negative bacteremias related to the Enterobacterales are frequently caused by E. coli , K. pneumoniae , Enterobacter spp., and P. mirabilis . Infections acquired in the hospital are likely to be caused by members of antibiotic-resistant genera, such as Citrobacter , Enterobacter , and Serratia . Enterobacterales associated with diarrhea include Shigella spp., Salmonella spp., E. coli (enterohemorrhagic [Shiga toxin producing], enterotoxigenic, enteroinvasive, enteropathogenic, enteroaggregative), and Yersinia spp. Shigella spp. are rarely isolated from sources other than the GI tract, whereas Salmonella spp. are more likely to be isolated from other sources, such as urine or blood. Plesiomonas spp. have been added to the Enterobacterales and remain as the only oxidase-positive member of this family. They also can be associated with infections of the GI tract. Klebsiella granulomatis is the causative agent of donovanosis or granuloma inguinale, a disease characterized by chronic genital ulcers ( ).
Endotoxins that are present within the cell walls of the Enterobacterales, as well as other gram-negative bacilli, are responsible for much of the morbidity and mortality resulting from infections associated with these bacteria. Endotoxins consist of lipid and polysaccharide moieties with small quantities of amino acids and are often referred to as lipopolysaccharides . Lipopolysaccharides may elicit fever, chills, hypotension, granulocytosis, thrombocytopenia, disseminated intravascular coagulation, and activation of both classic and alternate complement pathways. Endotoxic shock is the result of gram-negative septicemia, with endotoxin reacting with macrophages, leukocytes, platelets, complement, and other serum proteins to increase the blood levels of proteolytic enzymes and vasoactive substances, resulting in pooling of blood, increased peripheral vasoconstriction, and diminution in cardiac output. It has become clear that the lethal effects of endotoxin are dependent on macrophage activation and responsiveness and that the production of cachectin from the activated macrophage plays a major role in causing profound shock and multiple-organ injury ( ).
Other pathogenetic factors of the Enterobacterales include the K1 antigen, which is associated with a high percentage of strains of E. coli , causing neonatal meningitis; the capsule of K. pneumoniae , which, like that of the pneumococcus, inhibits phagocytosis; the Vi antigen of Salmonella serotype typhi , which may interfere with intracellular killing of this organism; and various surface antigens, such as fimbriae, that mediate adherence of the organism to mucosal surfaces.
Plasmid-mediated factors appear to play an important role in the invasive properties of Salmonella , Shigella , and enteroinvasive strains of E. coli . Moreover, the heat-labile enterotoxins (LT) and the heat-stable enterotoxins of E. coli are plasmid mediated. LT stimulates adenylate cyclase in mucosal cells of the small intestine, which, in turn, activates cyclic adenosine monophosphate (cAMP). This causes secretion of fluid and electrolytes into the intestinal lumen, producing watery diarrhea. In contrast, heat-stable (ST) enterotoxins appear to activate guanylate cyclase.
The isolation of gram-negative bacilli from contaminated specimens is greatly facilitated by the use of differential and selective media ( Table 57.5 ). Eosin methylene blue (EMB) and MacConkey agar can be used interchangeably as minimally selective and differential media to initially select for and differentiate lactose-fermenting from non–lactose-fermenting gram-negative bacilli. XLD and HE agars are more selective differential media that are especially useful in selecting for Salmonella spp. and Shigella spp. in heavily contaminated specimens such as stool. Bismuth sulfite is a highly selective medium that is especially useful for the detection of salmonellae in endemics or epidemics. Salmonella spp. produce H 2 S and are easily recognized by the production of colonies with black centers on XLD, HE, and bismuth sulfite agars. An enrichment medium, such as selenite-F or gram-negative (GN) broth, should be used to allow detection of low numbers of Salmonella spp. and Shigella spp. in stool. Cefsulodin-irgasan-novobiocin (CIN) agar incubated at room temperature is selective and differential for the recovery of Yersinia enterocolitica . Colonies will appear as bull’s-eyes with red centers and transparent borders. MacConkey agar with sorbitol as the fermentable sugar is a differential medium that is capable of differentiating the sorbitol-fermentation–negative E. coli 0157:H7, which is associated with hemolytic uremic syndrome from most other E. coli ( ).
| Medium | Gram-positive Bacteriostatic Agent | Fermentable Carbohydrate | Indicator | Colony Color Fermenter | Nonfermenter | Category |
|---|---|---|---|---|---|---|
| Eosin methylene blue (EMB) | Eosin Y Methylene blue |
Lactose ∗ | Eosin Y Methylene blue |
Red or black with sheen | Colorless | S, D |
| MacConkey | Crystal violet Bile salts |
Lactose | Neutral red | Red | Colorless | S, D |
| Xylose-lysine-deoxycholate (XLD) | Bile salts | Xylose Lactose Sucrose |
Phenol red | Yellow | Red | S, D |
| Hektoen enteric (HE) | Bile salts | Salicin Lactose Sucrose |
Bromthymol blue | Yellow-orange | Green, blue-green | S, D |
| Salmonella shigella (SS) | Bile salts | Lactose | Neutral red | Red | Colorless | S |
| Bismuth sulfite (BS) | Brilliant green | Glucose | Bismuth sulfite | † | † | S |
| Thiosulfate citrate bile salts sucrose (TCBS) ‡ | Bile salts pH 8.6 | Sucrose | Thymol blue Bromthymol blue |
Yellow | Colorless | S, D |
Multiple commercially available nucleic acid amplification tests can be used to detect bacterial stool pathogens ( ; ). These assays can be more sensitive and provide results in a more timely manner than culture. Since public health departments often require stool pathogen isolates to be submitted for outbreak investigations, discussions with all parties should occur before bringing on one of these assays. In addition, the ability to culture bacterial stool pathogens should still be retained in case susceptibility testing is needed.
For some of the Enterobacterales, a few simple colonial characteristics or biochemical reactions can be used to presumptively identify an isolate. For example, Proteus spp. swarm on blood agar, Klebsiella spp. form lactose-positive mucoid colonies, Serratia marcescens may produce a red pigment, Salmonella spp. produce H 2 S, and E. coli is indole positive. Definitive identification of these and other species requires additional biochemical testing and/or molecular methods. Innumerable schemes based on the use of conventional biochemical media have been described for identification of the Enterobacterales. Some differential characteristics of the Enterobacterales most commonly encountered in the clinical laboratory are shown in Table 57.6 . Commercially prepared kits and automated devices are available that offer convenience and accuracy in identifying the vast majority of isolates belonging to the Enterobacterales. In some instances, identification is accurately made in a few hours. Semiautomated systems may combine identification and antimicrobial susceptibility testing in a single disposable unit. In general, accuracy of identification among these systems is very high and comparable. MALDI-TOF has been shown to be an excellent tool for the identification of members of the Enterobacterales with the exception of Shigella spp. and can usually provide results more quickly than traditional or semiautomated biochemical methods ( ).
| Test | Escherichia coli | Klebsiella pneumoniae | Klebsiella oxytoca | Proteus mirabilis | Proteus vulgaris | Shigella spp. | Citrobacter freundii | Yersinia enterocolitica | Enterobacter cloacae | Serratia marcescens | Morganella morganii | Providencia alcalifaciens | Salmonella Serotype choleraesuis | Salmonella Serotype typhi | Salmonella Serotype paratyphi A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Indole | + | – | + | – | + | D | – | D | – | – | + | + | – | – | – |
| H 2 S (in triple sugar iron agar) | – | – | – | + | + | – | + | – | – | – | – | – | D | + | – or + |
| Urease | – | + | + | + | + | – | D | + | D | – or + | + | – | – | – | – |
| Motility | + or – | – | – | + ∗ | + ∗ | – | + | + + (25C)/– (37°C) | + | + | + or – | + | + | + | + |
| Acid produced from lactose | + | + | + | – | – | – | + or (+) | D | D | – | – | – | – | – | – |
There are two species of Salmonella , S. enterica and S. bongori (rarely isolated from humans), each of which contains multiple subspecies. Six subspecies of S. enterica are known, with subspecies I ( S. enterica subsp. enterica ) being the most common human isolate. More than 2000 serotypes of Salmonella have been identified, most belonging in the subspecies enterica . Serotyping is based on immunologic reactivity of the heat-stable somatic “O” antigens, which are predominantly lipopolysaccharide in content, and the heat-labile flagellar “H” antigens. In the United States, S. serotypes typhimurium and enteritidis are the most common. S. serotype typhi also produces a heat-labile capsular polysaccharide Vi antigen. In practice, most clinical laboratories identify isolates as Salmonella spp. based on biochemical reactions and use group-specific immunologic reagents to assign isolates to a specific serogroup. Commercial slide agglutination tests to differentiate large serogroups—A, B, C, and D—are useful in differentiating typhoidal salmonella from nontyphoidal strains. S. serotype typhi carries the D serogroup and Vi antigen. Further identification of the specific serotype is generally performed only by state health departments or other reference laboratories ( ).
Isolates biochemically resembling Shigella are also classified by the reactivity of the “O” antigen, as are isolates of E. coli that are identified as potential causes of diarrhea and hemolytic uremic syndrome. Such E. coli are classified by the type of toxin produced as well. Commercial kits are available to identify E. coli O157 and to detect some types of toxins in culture or stool specimens. However, this type of testing can be sent out to referral laboratories or state health departments. The CDC has recommended that all laboratories consider testing stool samples for the presence of Shiga toxins I and II produced by certain strains of enterohemorrhagic E. coli (O157:H7 and other serotypes). Shiga toxin is also produced by Shigella , but by the species S. dysenteriae , which is not often seen in the United States. Antigen tests are available and are widely used in clinical laboratories for this toxin ( ).
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here