Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
The authors thank members of the medical genetics staff who worked tirelessly to help accumulate the clinical pictures and the families who graciously consented to allow photographs to be taken to advance teaching and education.
The field of pediatric genetics and dysmorphology is complex, interesting, and rapidly evolving. Our knowledge base is gleaned from the careful observations of master clinicians and scientists who recognized clinical characteristics and patterns of malformation in individuals with genetic, teratogenic, developmental, and metabolic problems. They have provided us with a framework for the investigation of patients from clinical and laboratory perspectives. In addition to classic cytogenetics, molecular cytogenetics methods have been increasingly incorporated in clinical settings and have greatly assisted evaluation, enabling far greater understanding of the molecular and physiologic basis of these disorders, and have greatly increased the rate of diagnosis of children with genetic and metabolic disorders. However, even with the availability of an ever-widening array of confirmatory tests, clinical evaluation of patients remains an essential component of the complete assessment of children and adults with genetic diseases and dysmorphic conditions. This stems from the fact that careful evaluation can substantially reduce the number of differential diagnostic possibilities and, thereby, the number of diagnostic tests and the total expense.
Visual identification of dysmorphic features, baseline anthropometrics combined with serial measurements with recognition of patterns of malformation and behavioral phenotypes, remains an integral part of the diagnostic algorithm. As in pediatrics in general, genetic disorders should be investigated on the basis of a careful history, with a family pedigree and a thorough physical examination including evaluation for the presence of major and minor anomalies, and thoughtful laboratory testing. This chapter is designed to present clinicians who care for children with background on the general principles of genetics and dysmorphology, as well as updated information about important advances in our field. Although not exhaustive, it provides a framework for the broad categories of genetic diseases and discusses an approach to the evaluation of the dysmorphic child. Definitions and examples of the types of disorders resulting in genetic and/or congenital anomalies in children are described, including malformations, deformations, disruptions, associations, and sequences. We include examples of disorders inherited through classic mendelian inheritance patterns, including single-gene mutations, such as Marfan syndrome, or de Lange syndrome, metabolic disorders such as phenylketonuria (PKU) or Smith-Lemli-Opitz syndrome, as well as examples of nonmendelian disorders, such as teratogenic exposures in utero and disruptions or deformations of previously normal fetal structures. Etiologic mechanisms of diseases, such as imprinting abnormalities, expansions of trinucleotide repeats in nuclear deoxyribonucleic acid (DNA), mitochondrial DNA, whole exome and genome sequencing, as well as the fields of -omics (metabolomics, proteomics, etc.), are introduced. As testing options grow, there is acknowledgement of the expanding role of genotype to phenotype correlation and its importance in the genomic era.
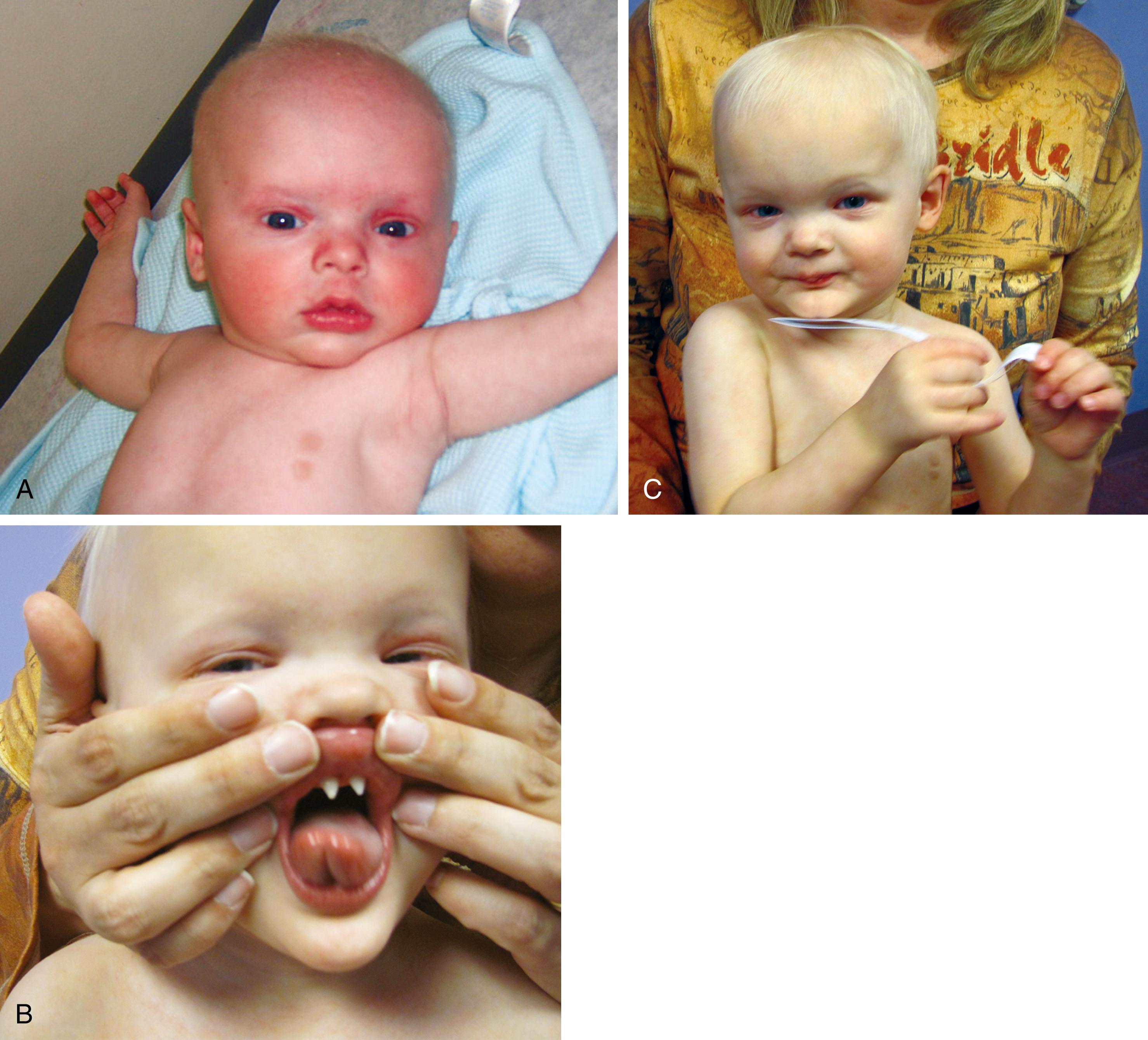
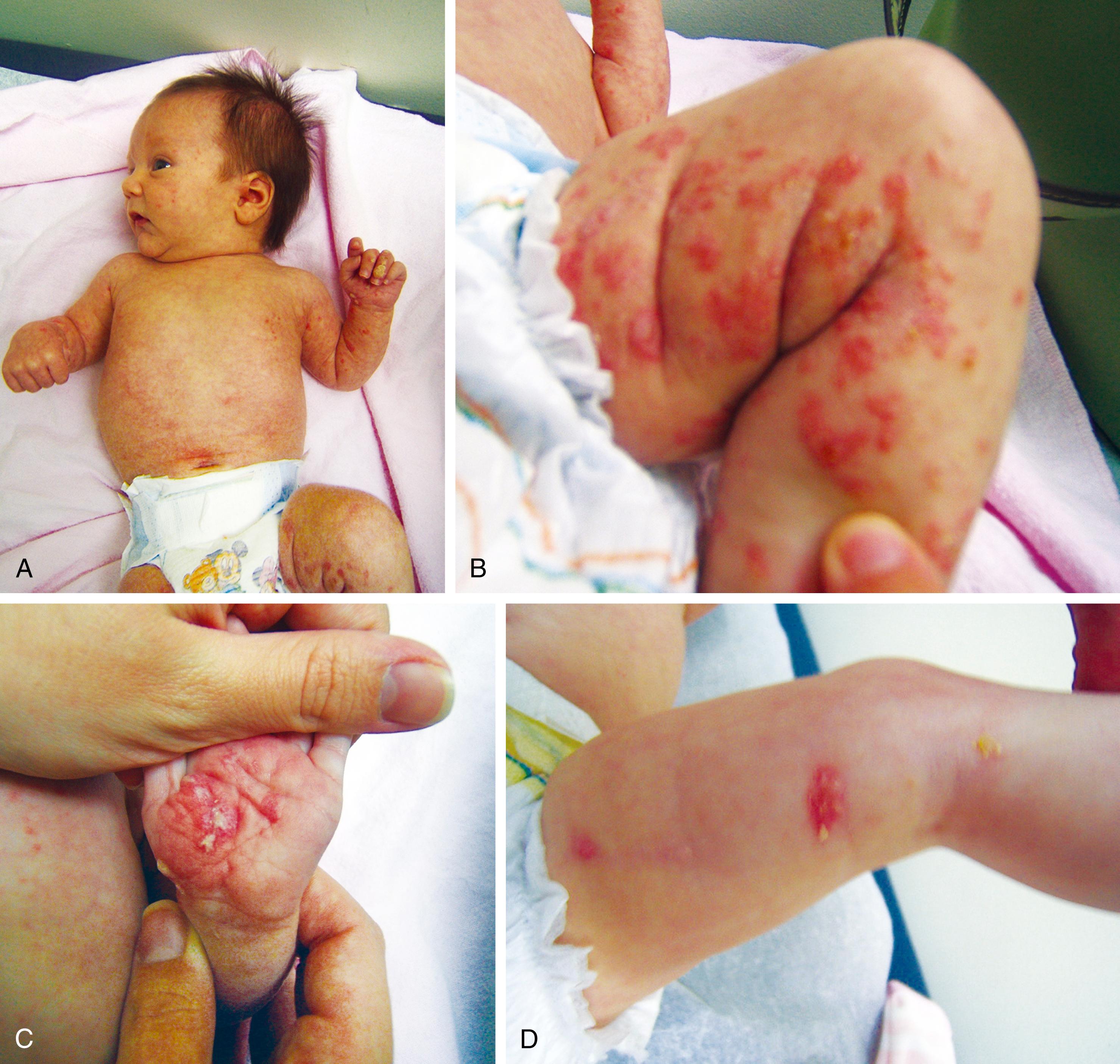
Approximately 2% to 3% of liveborn infants have an observable physical structural abnormality ( major anomaly). This number rises to about 4% to 5% by the time the child is old enough to attend school. Structural differences can be determined to be either major or minor in character ( Table 1.1 ; Figs. 1.1 and 1.2 ). Major structural anomalies have functional significance. Examples are polydactyly, colobomas of the iris (see Chapter 20 ), meningomyelocele, and cleft lip. Minor anomalies are usually of cosmetic importance only. Examples are epicanthal folds of the eyes, single transverse palmar creases, and supernumerary nipples. The incidence of isolated major anomalies in the general newborn population is approximately 1%, and the incidence of minor anomalies is approximately 14%. Both are more common in premature newborns.
| Category | Major | Minor |
|---|---|---|
| Craniofacial | Choanal atresia | Plagiocephaly |
| Flat occiput | ||
| Eyes | Coloboma of iris | Epicanthal folds |
| Ears | Microtia | Preauricular pit |
| Hands | Polydactyly Absent thumbs |
Single transverse palmar crease |
| Clinodactyly |
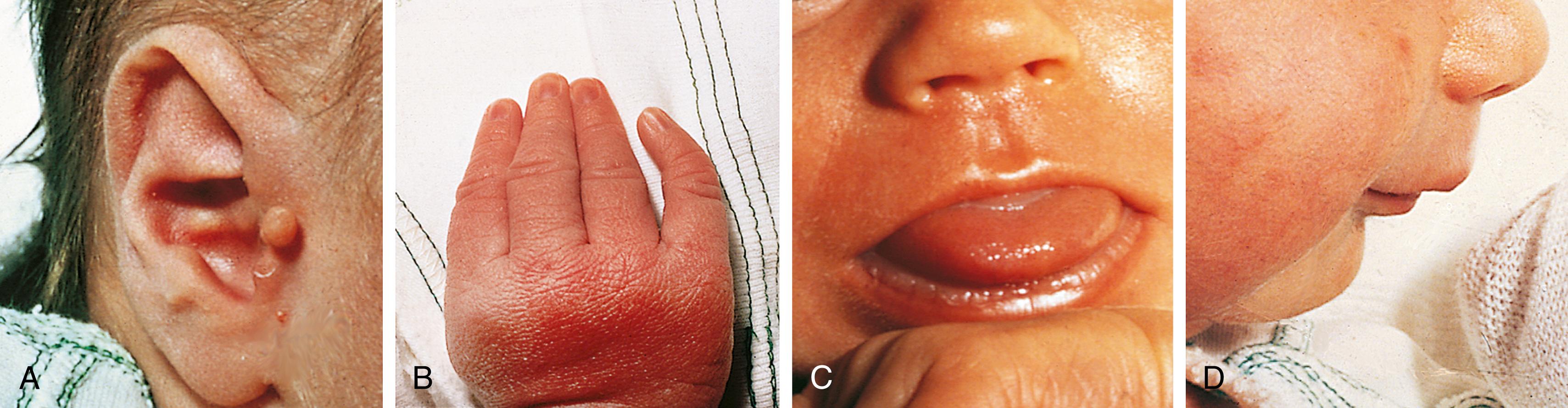
The probability of an infant having a major anomaly increases with the number of minor anomalies found. Thus all children with multiple minor anomalies warrant a careful clinical assessment in order to find potentially significant occult major anomalies. Once an anomaly is identified, assessing its significance begins with a determination of whether the anomaly in question is a single localized error in morphogenesis or one component of a multiple malformation syndrome. An understanding of the pathophysiologic mechanisms that produce structural abnormalities or differences provides an opportunity to define the types of structural abnormalities seen. This also assists the process of identifying the etiology and arriving at a specific diagnosis, which then can be useful in determining the prognosis and estimating the risk of recurrence of a similar problem in future pregnancies.
Definitions of the classifications of structural anomalies aid in communication between clinicians and in the process of evaluation and are summarized below:
Malformation: A malformation is an abnormality of embryonic morphogenesis of tissue. It usually results from genetic, chromosomal, or teratogenic influences, but it can be of multifactorial etiology. Malformations are divided into two main categories: (1) those that constitute a single primary defect in development and (2) those that represent a single component of a multiple malformation syndrome. A multiple malformation syndrome can be defined as one having several observed structural defects in development involving multiple organ systems that share the same known or presumed etiology. Malformations often require surgical intervention.
Deformation: A deformation represents an alteration (often molding) of an intrinsically normal tissue caused by exposure to unusual extrinsic forces. A classic example is clubfoot, which may be the result of uterine constraint from crowding associated with a multiple gestation. A more severe example is the compressed facial features (“Potter facies”) of a child exposed to severe uterine constraint associated with oligohydramnios, due to renal agenesis (see Chapter 14 ). The vast majority of deformations respond to medical therapy alone and have a relatively good prognosis in contrast to malformations, which frequently require surgical intervention.
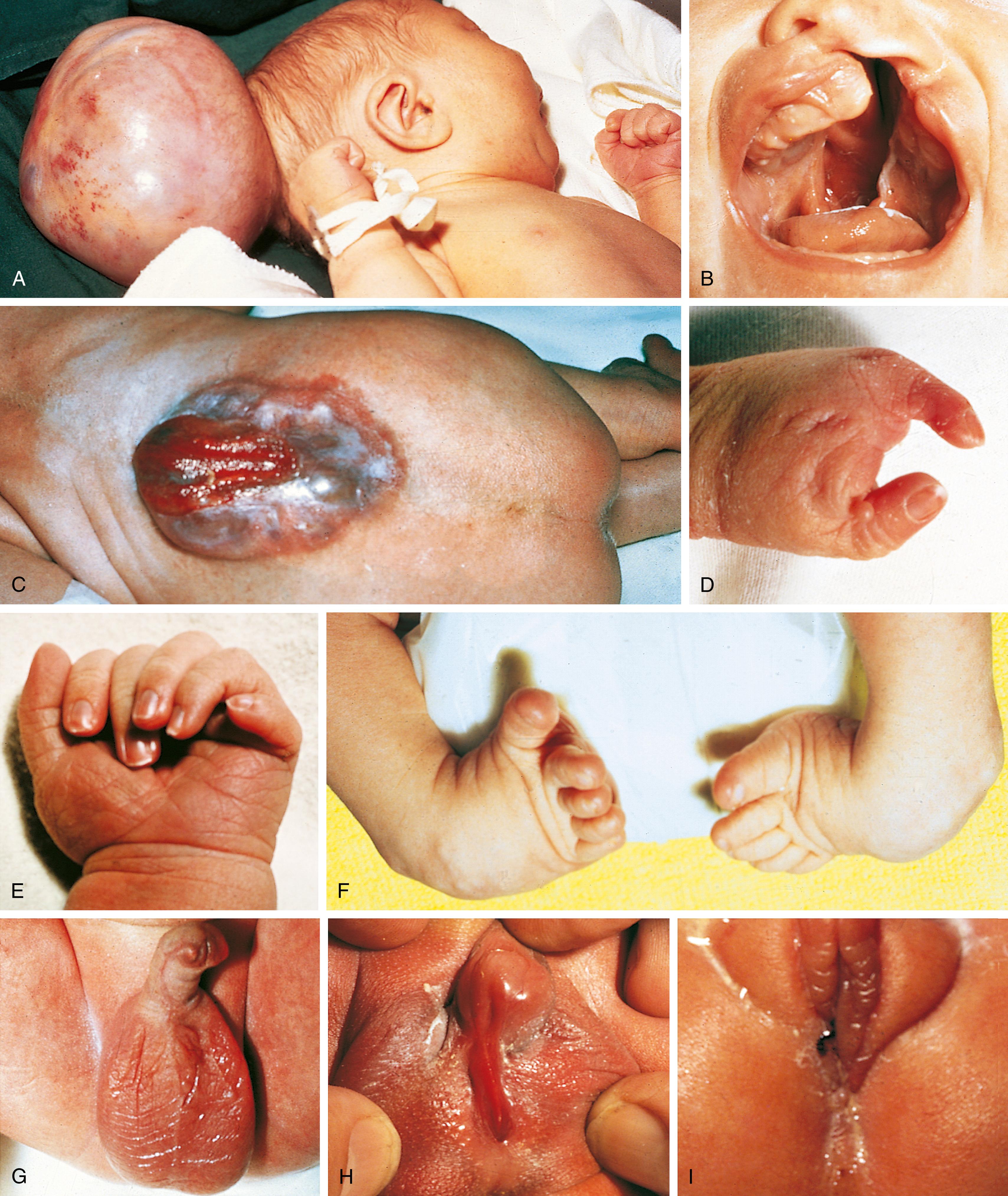
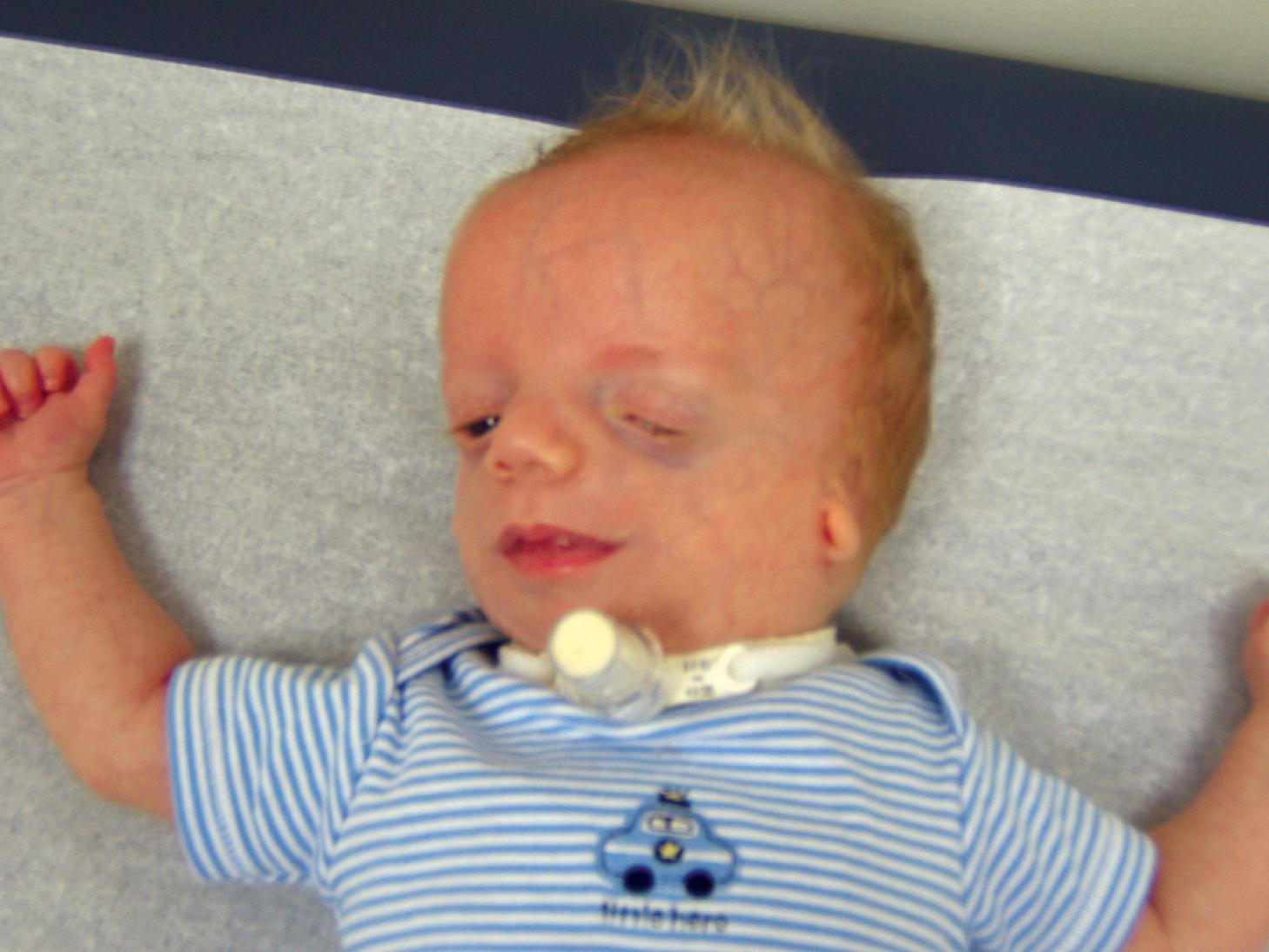
Disruption: A disruption represents a breakdown of normally formed tissue; the breakdown may be the result of vascular accidents or exposure to adverse mechanical forces that are usually more severe than those that produce deformation. A classic example is the combination of clefting, constriction bands, and limb reduction defects associated with the presence of amniotic bands (see Chapter 2 ). The earlier these vascular accidents or abnormal forces occur during embryogenesis, the more severe the resulting defects ( Fig. 1.3 ).
Dysplasia: Dysplasia is characterized by abnormal organization of cells within tissue, which usually has a genetic basis. An example is achondroplasia, the most frequent form of skeletal dysplasia.
( Note: Each of the preceding categories can have a sequence associated with it.)
Sequence: The term sequence refers to a recognizable pattern of multiple anomalies that occurs when a single problem in morphogenesis cascades, resulting in secondary and tertiary errors in morphogenesis and a corresponding series of structural alterations. A classic example is the Robin malformation or Pierre Robin sequence, in which the single primary malformation is microretrognathia (see Chapter 23 ). The resulting glossoptosis, or posterior placement of the tongue in the oropharynx, interferes with normal palatal closure if the lingual displacement occurs before 9 weeks’ gestation. The resulting cleft palate is U -shaped, rather than having the V shape that is usually seen in classic cleft palate, a finding that aids in recognition.
Association: An association is a pattern of malformations that occurs together too frequently to be due to random chance alone but for which no specific etiology is yet recognized.
The approach to the evaluation of a child with a dysmorphic abnormality is similar to a careful diagnostic evaluation of most pediatric problems, starting with a complete history and careful physical examination. In obtaining these, it is helpful to remember that there are six broad etiologic categories to be considered in the differential diagnosis: (1) a known syndrome, (2) an unknown syndrome, (3) a chromosomal abnormality, (4) a teratogen, (5) a congenital infection, and (6) a maternal disease and/or placental abnormalities.
The history should include the following:
Course of the pregnancy, complications including possible infections or environmental exposures, medications/substance abuse
Prior pregnancies, spontaneous abortions, stillbirths, or infant/child deaths for this couple
Labor/delivery/perinatal problems
Past medical history
Growth and development
Meticulous family history with family tree going back three generations and including the following:
Familial traits and growth characteristics
Familial physical or developmental disorders
Spontaneous abortions, stillborns, infant/child deaths in extended family
The physical examination entails the following:
Thorough general examination
A search for major and/or minor anomalies
Neurodevelopmental assessment
In addition, focused examination of immediate family members for physical characteristics and growth parameters and review of family photo albums may be helpful.
Determining how the child fits into the norms for growth and development for the general population, for the family’s ethnic group(s), and for the extended family is important. One continuing challenge is to determine whether the norms for the family are truly in the normal range for the general population and ethnic background or, in fact, constitute variability of a genetic trait present in its severe expression in the child or family member seen for evaluation.
The identification of a recognizable pattern of both major and minor anomalies provides the clinical dysmorphologist with a diagnosis, or a short list of differential diagnostic possibilities. Thus the detection of major and minor anomalies is critical in the diagnostic process. Identification of specific and unusual malformations that are uncommon and occur in only a few syndromes can be especially helpful. For example, finding that a child has long palpebral fissure length and pronounced fingertip fat pad size in combination with the pattern of anomalies typical of the Kabuki syndrome makes it extremely likely that the diagnosis is the Kabuki syndrome. Training in dysmorphology emphasizes the recognition of key components in patterns of malformation, as well as the specific findings useful in distinguishing syndromes with similarities from one another. Texts that outline currently recognized patterns of malformations can be helpful in assisting the clinician in the identification of specific features that can rule a diagnosis in or out. Commercial computer-based programs exist for syndrome identification; however, these are often more effectively used by experts in the field because of the complexity of terminology and the need for exacting descriptions of the anomalies present in a given child.
Cytogenetic and/or molecular studies should be performed on each child with a syndrome of congenital anomalies. Such studies may establish or confirm the diagnosis of an underlying genetic disorder and its hereditary potential and may possibly help map the chromosomal location or candidate genes for those syndromes known to be simple mendelian disorders.
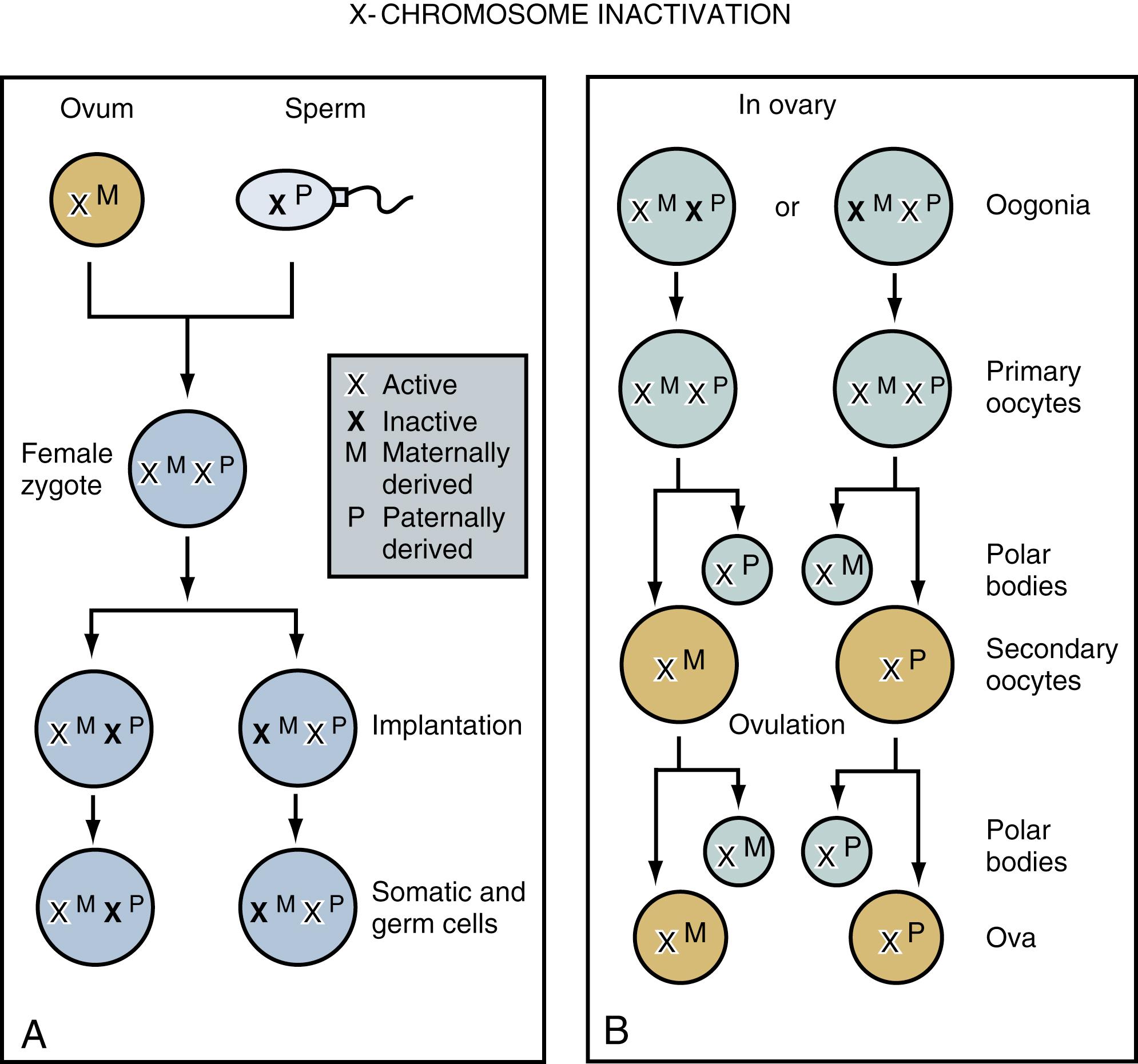
Productive insights gleaned from the results of the Human Genome Project have dramatically changed some of our understanding of how the human genome functions. Human hereditary factors are located in genes (the genome ). Approximately 10% are genes that encode proteins that are assembled to form tissue structures or to form enzymes that catalyze chemical reactions within cells. The other 90% have functions that are currently not clear (see also The Nature of Genes and Single-Gene Disorders, later). The genes are composed of DNA and are stored in intranuclear cell organelles called chromosomes. Each chromosome contains one linear DNA molecule folded over onto itself several times, as well as ribonucleic acid (RNA) and proteins. Because all genes exist in pairs, all chromosomes must likewise exist in pairs. The members of each pair of genes are called alleles, and the members of each pair of chromosomes are known as homologues. The conventional depiction of the constitution of homologues in the nucleus is called the cell’s karyotype ( Fig. 1.4 ). If at any gene locus the alleles are identical, that gene locus is homozygous. If the alleles are not identical, the gene locus is heterozygous.

Except for gametes, normal human cells contain 23 pairs of chromosomes, 46 in all. One of these pairs is concerned in part with inducing the primary sex of the embryonic gonads. These sex chromosomes are called the X and Y chromosomes, and they are not genetically homologous except in a few areas. Genotypic females have two X chromosomes, whereas genotypic males have an X and a Y chromosome. The remaining 22 pairs are called autosomes, and they determine non–sex-related (somatic) characteristics.
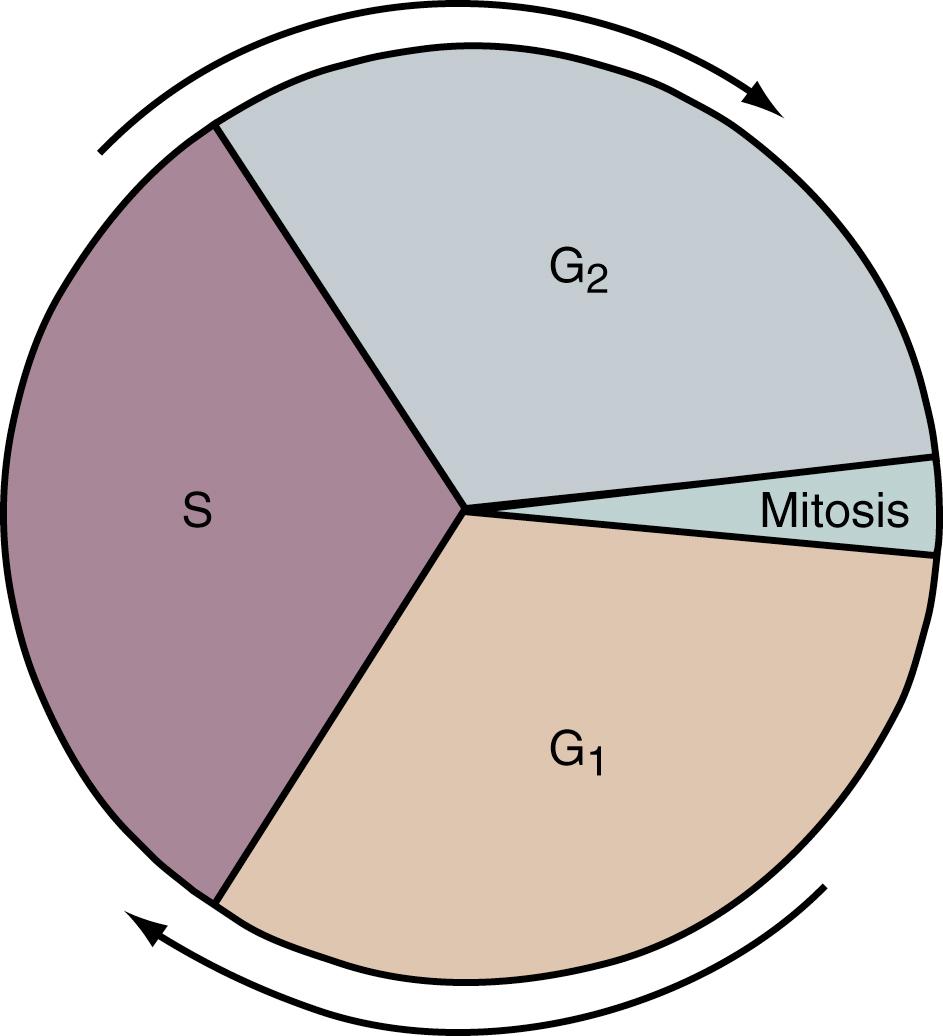
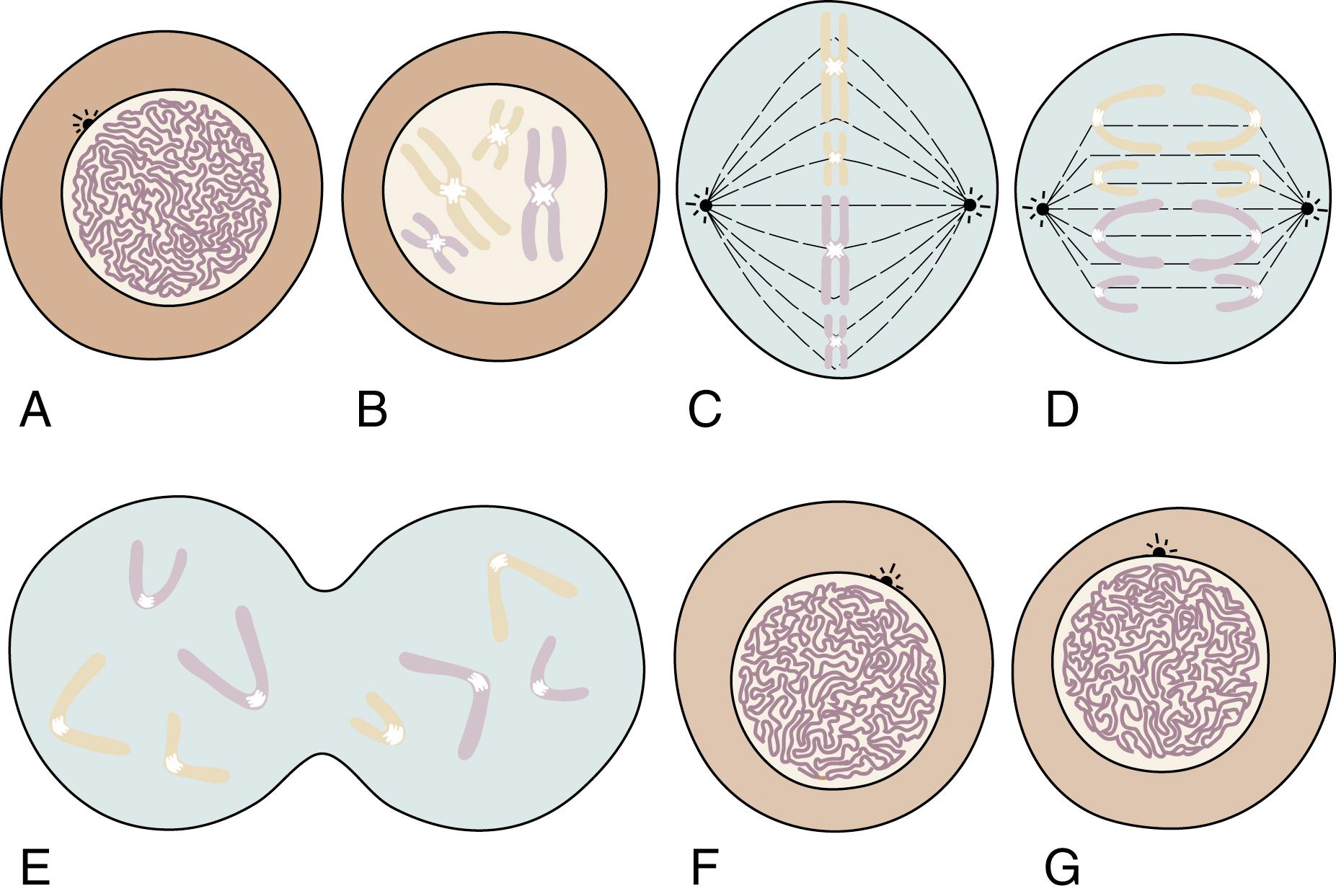
During most of a cell’s life cycle, chromosomes are diffusely spread throughout the nucleus and cannot be identified by morphologic means. Only when the cell divides does chromosome morphology become apparent ( Fig. 1.5 ). The in vitro life cycle and the cellular division, or mitosis, of a somatic cell are illustrated in Figs. 1.6 and 1.7 , respectively. The life cycle and divisions, or meiosis, of a germ cell are much more complex and are not suitable for ordinary clinical evaluation.
Any somatic cell that can divide in tissue culture can be used for chromosomal (cytogenetic) analyses for karyotyping. The most convenient tissue source is peripheral blood, from which lymphocytes can be stimulated to divide during 2 or 3 days of incubation in tissue culture medium. Fibroblasts obtained from skin remain a frequently used alternative when peripheral blood lymphocytes are not clinically suitable, but fibroblasts require an incubation period of 4 to 6 weeks. After death, lung tissue is the best tissue to culture for chromosomal analyses, followed by skin fibroblasts, although both require a 4- to 6-week incubation period. In urgent situations, preliminary chromosomal evaluation can be made within 4 to 24 hours.
Oftentimes the karyotype is supplemented within 48 to 72 hours by a molecular cytogenetics technique, either interphase or metaphase fluorescence in situ hybridization (FISH) and high-resolution molecular karyotyping using microarray-based comparative genomic hybridization (array-CGH). Array-CGH uses molecular probes to enable submicroscopic copy number changes at high resolution and has become standard of care as a first step in the investigation of patients with developmental delays, intellectual disability, and multiple congenital anomalies. This is an ever-evolving area, and pediatric clinicians are advised to discuss clinical and laboratory investigations with clinical geneticists and/or laboratory directors prior to sample collection.
Data from Hook (1992) suggest that upward of 50% of human conceptions terminate in a spontaneous abortion, typically so early during gestation that the pregnancy is never recognized. The earlier the abortion occurs, the more likely it is that the miscarried embryo had a chromosomal abnormality. Of recognized first-trimester abortuses, 50% are chromosomally abnormal, compared with 5% of later embryos. Among the chromosomally abnormal miscarried embryos, the most frequent abnormalities are triploidy (69 chromosomes), trisomy 16, and 45,X (Turner syndrome) ( Table 1.2 ). Generally speaking, triploidy and trisomy 16 are not compatible with life and are only rarely seen among liveborn infants. Despite the fact that Turner syndrome is relatively common among liveborn infants, the majority of conceptuses with 45,X also abort spontaneously. The incidence of chromosomal abnormalities among liveborn infants in general is about 6 in 1000, but for infants who are stillborn or who die in the immediate perinatal period, the number is increased to approximately 50 in 1000.
| Among Spontaneous Abortuses | Incidence |
|---|---|
| Overall incidence | 32.0% |
|
52.0% |
|
5.8% |
| Type of abnormality seen in spontaneous abortions | |
|
|
|
|
|
|
|
|
|
| Among Liveborns | Number of Cases Per 1000 |
|---|---|
| Overall incidence | 6.20 |
| Abnormality of autosomes | 4.19 (males and females) |
|
|
|
|
|
|
| Abnormality of sex chromosomes | 2.03 (males and females) |
|
|
|
a About one-quarter of all conceptuses are chromosomally abnormal. About 50 in 1000 stillborns have a chromosomal abnormality.
Aneuploidy refers to an abnormality in chromosome number, in humans a chromosome number different from an even multiple of 23 (the haploid number) ( Fig. 1.8 ). In aneuploidy, there are typically 45 or 47 chromosomes instead of the usual 46. Rarely, multiples of the X or Y chromosome result in individuals with 48 or 49 chromosomes.

If aneuploidy occurs in a gamete as a result of an error of chromosomal division (nondisjunction or anaphase lag) during meiosis, all cells are affected in the fertilized embryo. With subsequent pregnancies, the risk for another aneuploidy (same or different) is increased approximately 1% to 2% above the general population risk. The risk of nondisjunction increases independently with maternal age ( Table 1.3 ). Although the mechanism of this increased risk is unknown, preconception genetic counseling is recommended in those with a family history of aneuploidy, multiple miscarriages, or increasing maternal age.
| Anomaly | Frequency | Management |
|---|---|---|
CNS:
|
|
Supportive and symptomatic |
| Congenital heart disease: AVSD, endocardial cushion defect, VSD, vascular anomaly Adult onset of valvular prolapse |
45% | Cardiology involvement |
| Characteristic facial features: Up-slanting palpebral fissures, small nose with low nasal bridge, epicanthal folds, small external ears | Common | Supportive and symptomatic |
| Eyes: Brushfield spots (speckled irides), strabismus, lenticular opacities, refractive errors, blocked tear ducts, dental concerns |
Common | Symptomatic |
| Neck: Short with redundant fold |
Common | Detected in utero and postnatally. Neck stability checks due to C1–C2 |
| GI: Duodenal atresia, Hirschsprung disease, omphalocele, imperforate anus, cholelithiasis |
5%–12% | Surgical repair |
| Endocrine: Thyroid athyreosis, goiter, hyperthyroidism, primary gonadal deficiency, fertility |
Common | Early recognition and treatment Occasional pregnancy in female (approx. 30%). 3 males have had documented paternity. |
| Hematology/oncology leukemia, neonatal leukemoid reaction | Rare (1%) | |
| Musculoskeletal: Cervical spine instability, hypoplasia of pelvis, small hands with short fifth fingers and single palmar creases, wide gap between the first two toes of feet (sandal gap) |
Common | Supportive care and surgery as needed |
| Genitalia: hypogenitalism | Common | |
| Skin: Dry, hyperkeratosis, frequent infections, hair (fine and sparse, early greying) |
Common | |
| Allergy immunology (low T- and B-cell counts, small thymus, decreased antibody response to immunizations, decreased neutrophil chemotaxis) | Common |
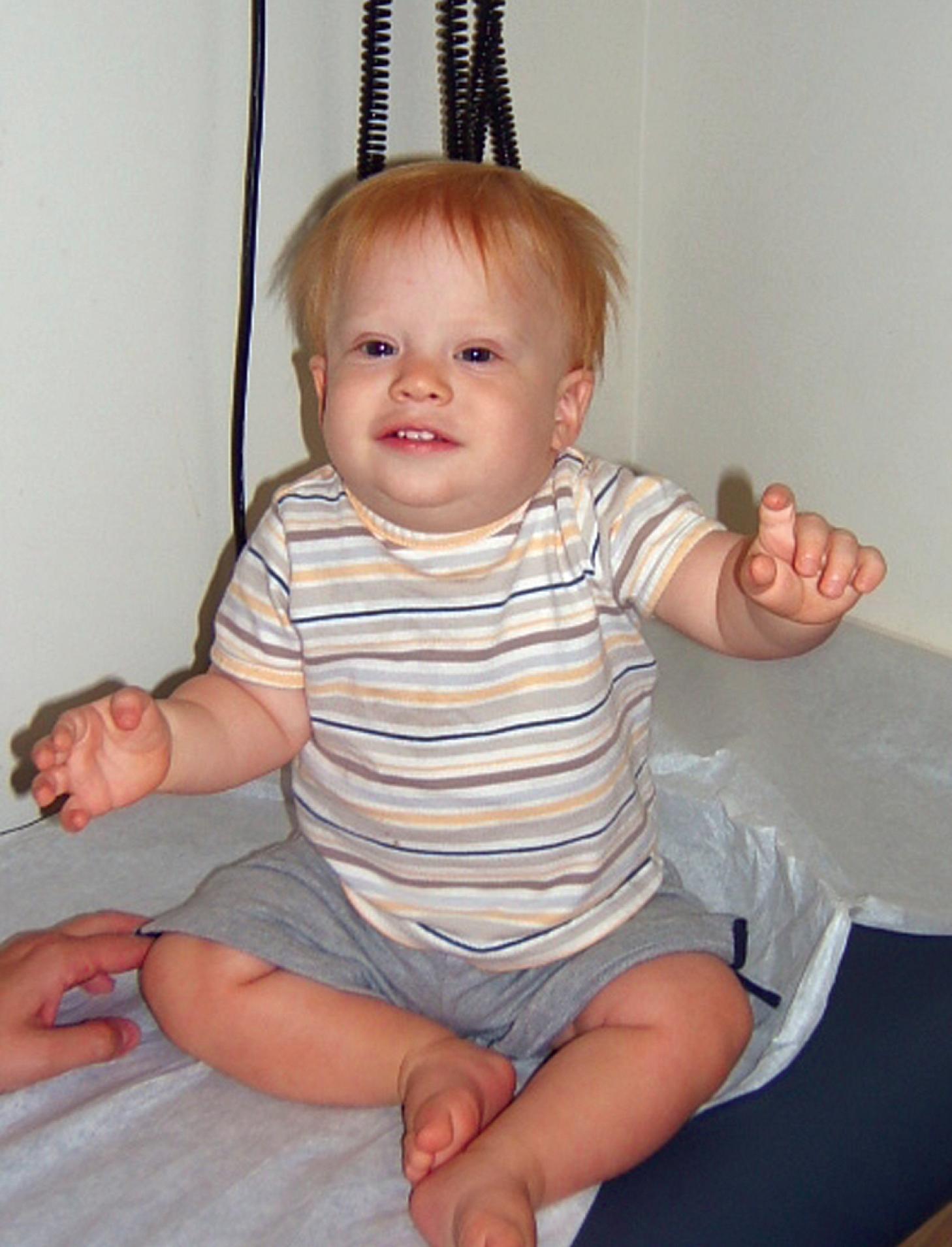
Mosaicism, the presence of two or more genetically different cell lines within an individual, can result from an error in division during either meiosis (preconception) or mitosis (post conception). When aneuploidy is present at conception, in some cases the extra chromosome is dropped from one or more cells during post-conception cell division, leading to mosaicism in the fetus for normal and aneuploid cell lines. In other cases of mosaicism the one-celled embryo (zygote) is chromosomally normal and a division error occurs after fertilization, during mitosis of an embryonic somatic cell, resulting in aneuploidy. Balanced and unbalanced chromosome aberrations and uniparental disomy (UPD) can be encountered in this setting. Generally speaking, if parents have a child with aneuploidy, their recurrence risk in future children is 1% to 2%; the reason for the increased recurrence risk is not fully understood but may reflect an undetectable aneuploid mosaic cell line in a parental gonad ( Fig. 1.9 ).
In some cases, chromosomal mosaicism can have visible effects. For example, in hypomelanosis of Ito the skin manifests marbleized or mottled areas of hypopigmented whorls along the Blaschko ( Fig. 1.10 ). Karyotyping from normal, hypo, or hyperpigmented regions can reveal mosaic chromosomal abnormalities in each region. In addition, individuals with hypomelanosis of Ito can have multiple congenital anomalies, dysmorphic features, variable intellectual disability, and other neurologic findings.
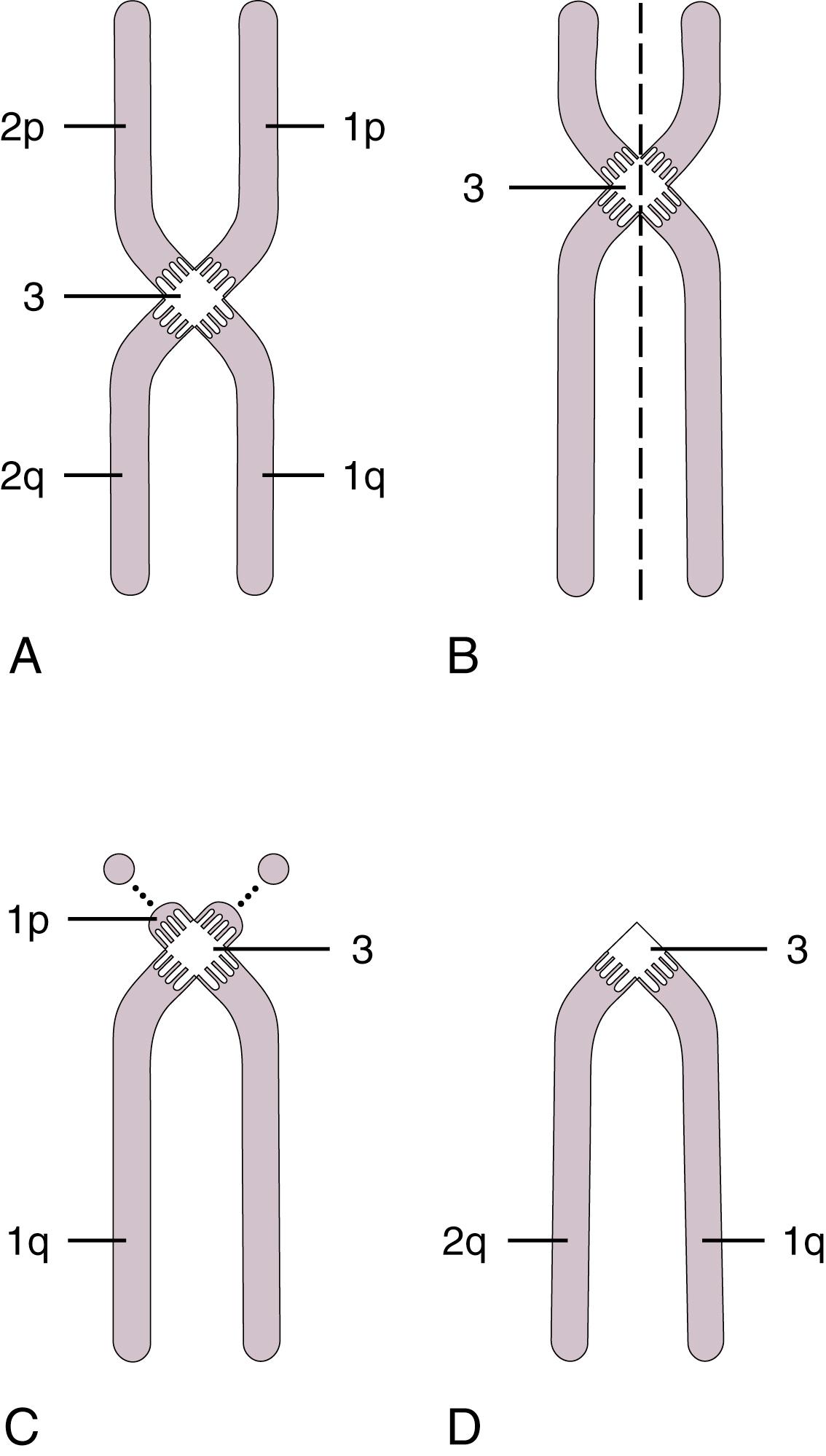
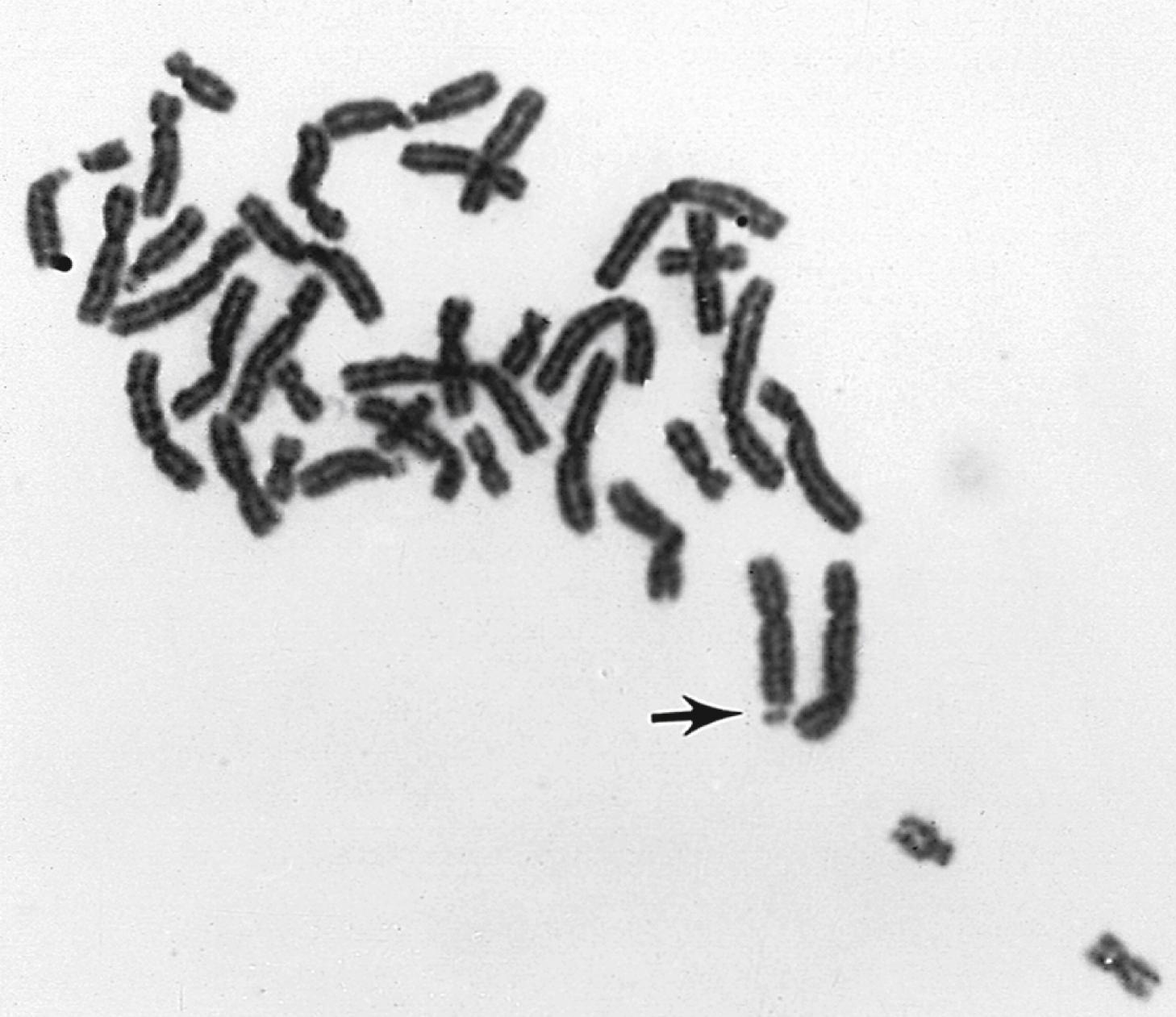
Chromosomes can be normal in number (diploid) but still be abnormal in structure. Inversions ( Fig. 1.11 ), deletions ( Fig. 1.12 ), and translocations ( Fig. 1.13 ) of genetic material are examples of structural chromosomal abnormalities. These can arise as new (sporadic) mutations in the egg or sperm from which the embryo was formed, in which case the parents’ recurrence risk for another child with a chromosomal abnormality is again 1% to 2%. However, the abnormality may also be inherited from a phenotypically normal parent who is a “carrier” of a balanced structural chromosomal abnormality ( Fig. 1.14 ). About 1 in 520 normal individuals carries a balanced but structurally abnormal set of chromosomes, called a chromosome translocation. The term balanced, for the purposes of this chapter, means that on cytogenetic analysis the structural abnormality does not appear to have resulted in any net loss or gain of genetic material, simply its relocation to a different place in the genome. A familial balanced translocation can be transmitted through multiple generations, but in some cases the translocated genetic material is passed down unevenly during mitosis, resulting in a fetus with aneuploidy.
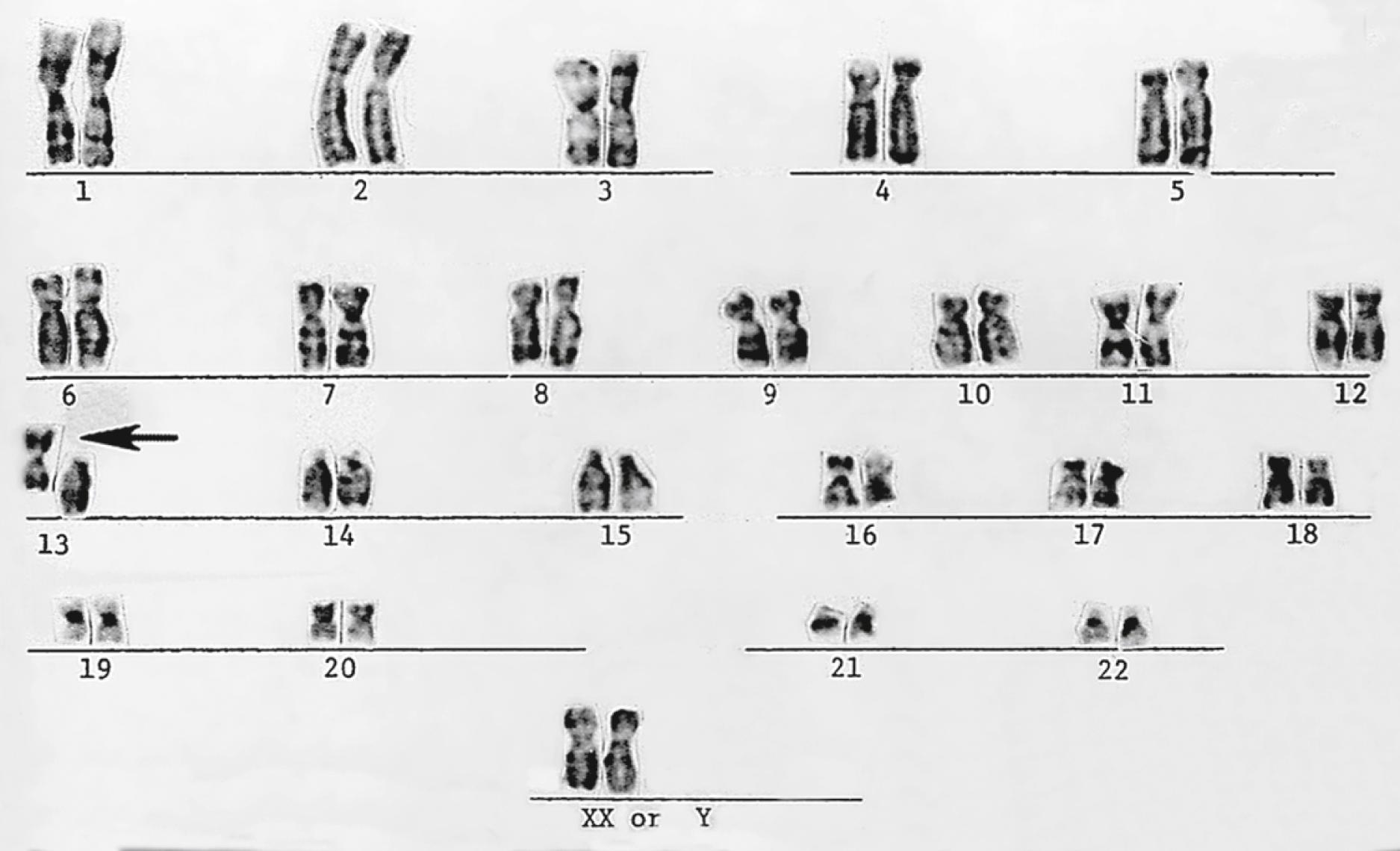
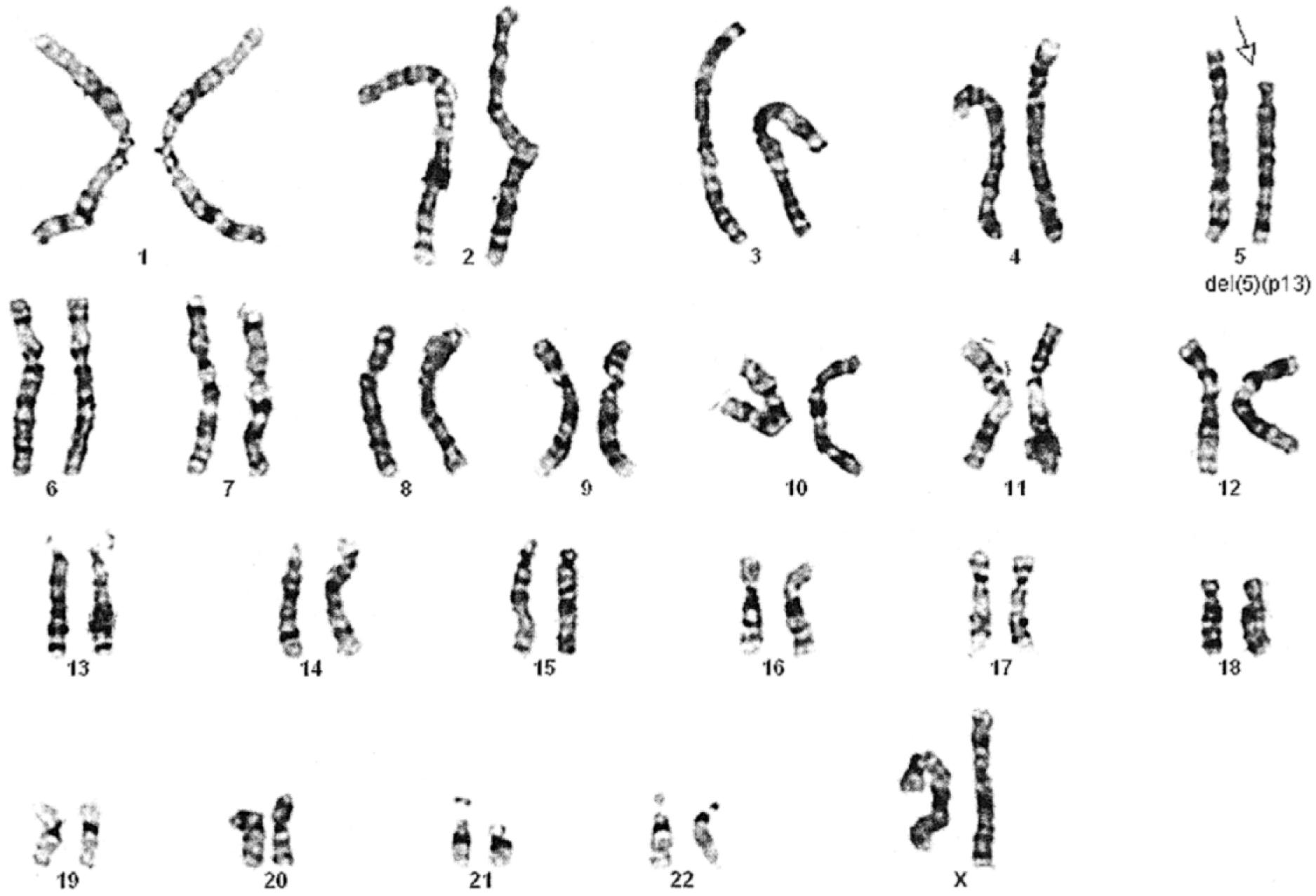
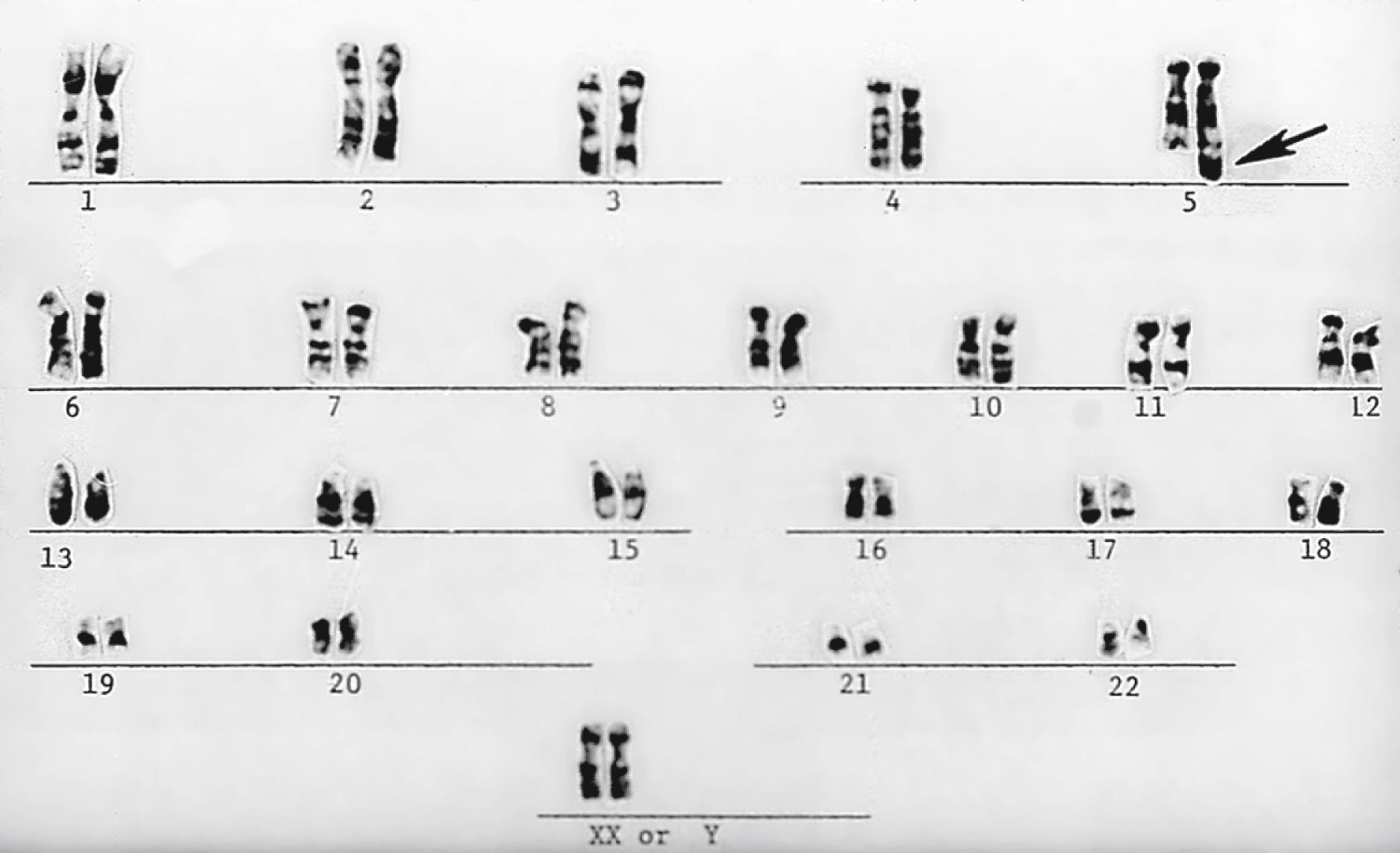
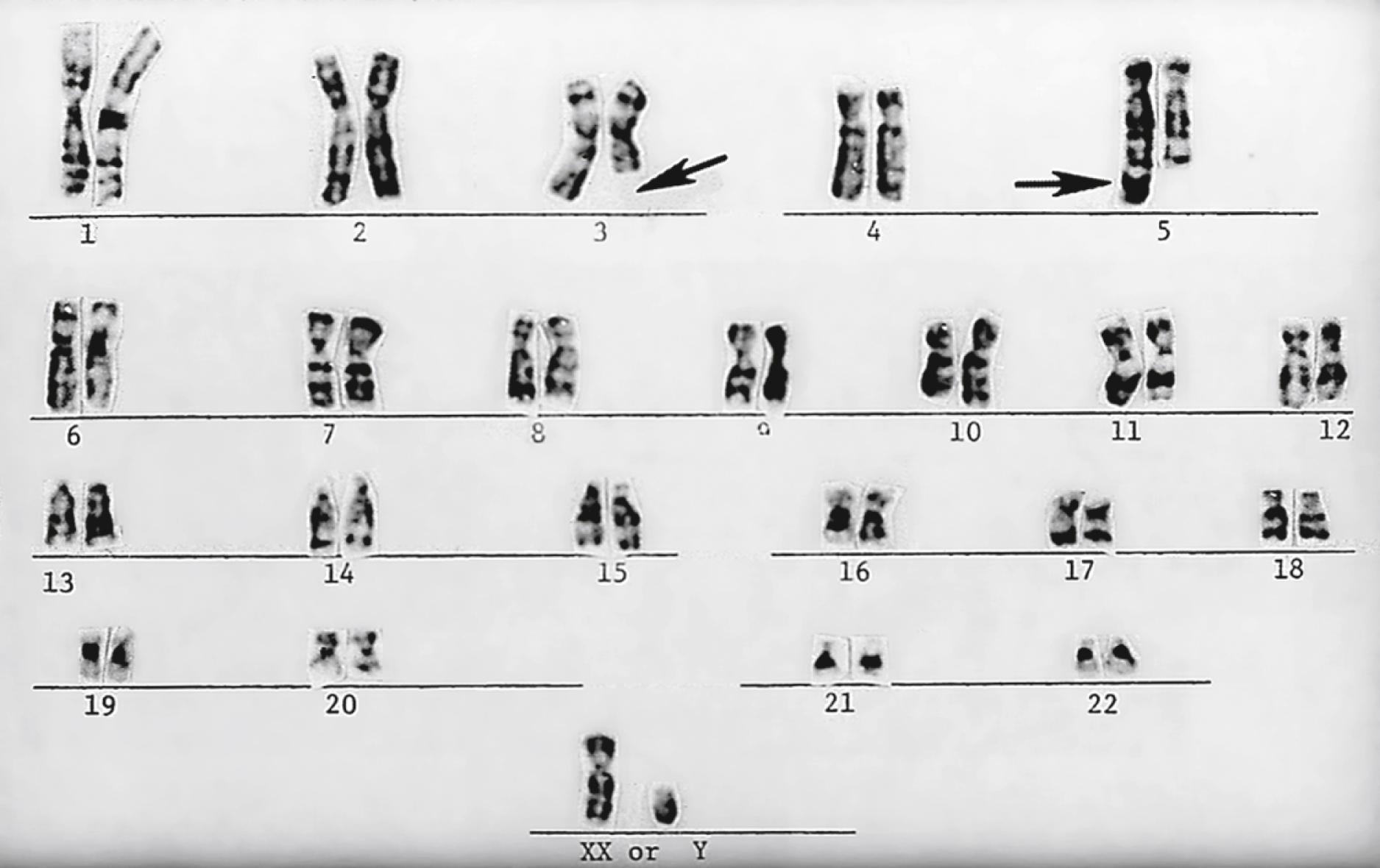
A frequent way in which families with apparently balanced chromosome translocations come to medical attention occurs when a child is born with structural malformations and on karyotyping is found to have an unbalanced chromosome translocation. Data suggest that a small percentage of individuals with de novo apparently “balanced” translocations are actually mildly affected clinically by variable degrees of cognitive and physical deficits. Thus high-resolution chromosome analyses and molecular cytogenetics techniques may be indicated, particularly when assessing new (de novo) translocations in a child. Parental karyotypes are used to distinguish the etiology and are crucial in providing accurate genetic counseling regarding future pregnancies for that couple.
Chromosomal abnormalities of either number or structure are likely to have a detrimental effect on the phenotype of an affected individual. Aneuploidy of an autosome, or non-sex chromosome, generally significantly impairs physical and cognitive development. However, aneuploidy of a sex chromosome may have little or no apparent effect on the phenotype. One should look for clustering of abnormalities in family members to suggest a problem, although their absence does not rule out a chromosomal abnormality.
Carriers of an inherited or a de novo reciprocal translocation are usually genetically balanced and are subsequently normal. However, their conceptuses can be genetically unbalanced and may abort spontaneously or be born with major congenital anomalies. A history of unexplained infertility, multiple spontaneous abortions (three or more), and particularly of a previous birth to the couple or to a close relative of a child with dysmorphic findings and/or major anomalies may be an indication that one of the parents carries a balanced chromosomal translocation or rearrangement. Thus a chromosome study on the couple is indicated, and if translocation is found, they should seek antenatal genetic counseling. This may also be advisable for extended family members. The risk for an individual carrying a balanced translocation to have a liveborn child with an unbalanced translocation varies from 4% to 20%, depending on the reproductive history of the individual and other family members who carry the balanced translocation.
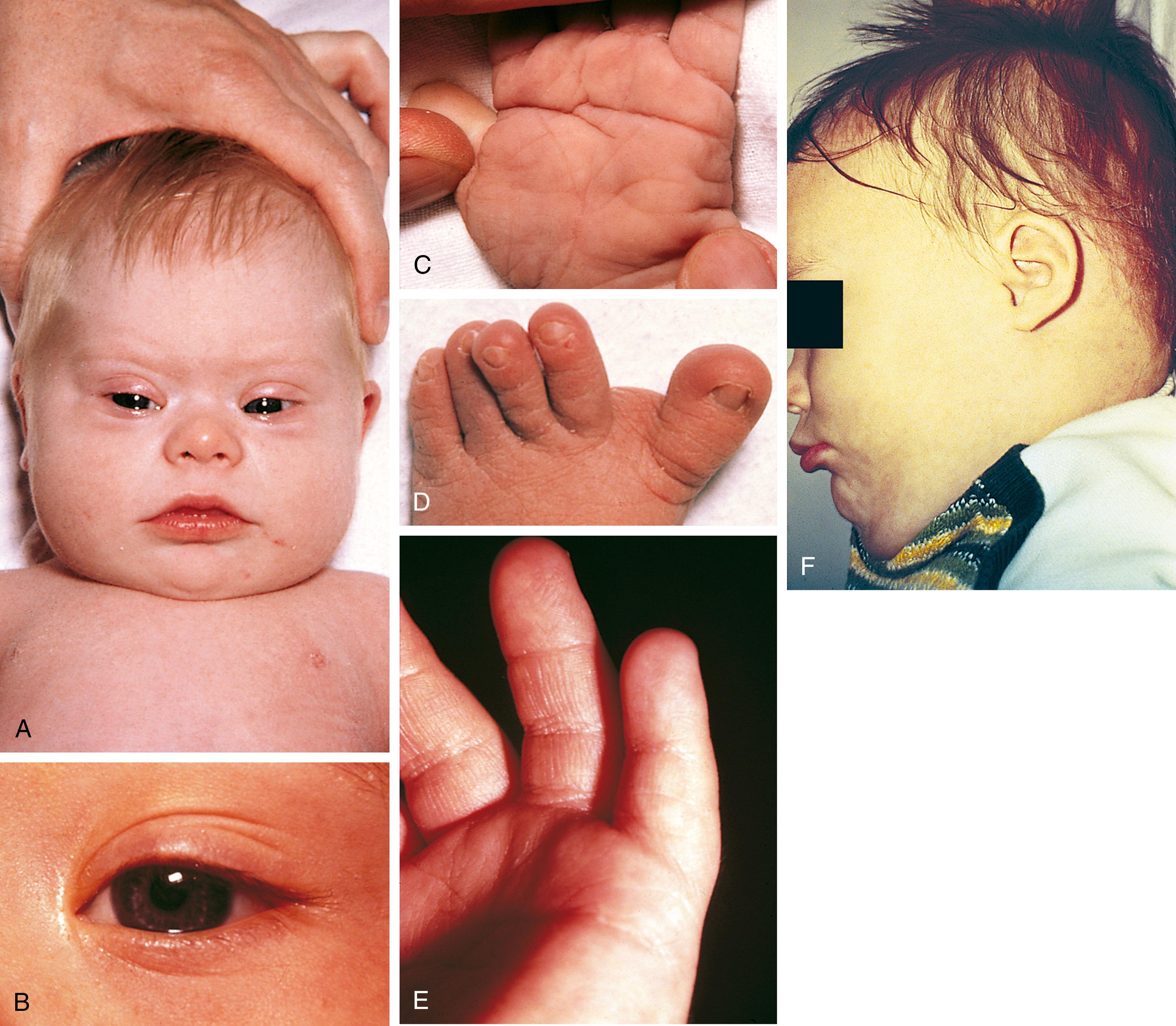
The worldwide incidence of Down syndrome among liveborn infants is approximately 1 in 660, with 45% of affected individuals born to women older than 35 years old. The incidence of Down syndrome among conceptuses is far greater than among liveborns because the majority of affected fetuses spontaneously abort. No single physical stigma of Down syndrome exists; rather, the clinical diagnosis rests on finding a recognizable constellation of clinical characteristics, including a combination of major and minor anomalies (see Figs. 1.9 and 1.15 , Table 1.3 ). The etiology of Down syndrome is trisomy 21, the presence of an extra chromosome 21 either as a simple trisomy or as part of a chromosome 21 fused (translocated) with another chromosome. Chromosome studies should therefore be performed on the parents and appropriate family members of an individual with translocation Down syndrome. Cases of mosaicism, in which trisomy 21 cell lines coexist with cell lines with the standard 46 chromosomes, exist as well and may range in phenotype from normal to that typical of complete trisomy 21. An association between trisomy 21 and advanced maternal age is clear ( Table 1.4 ).
| Maternal Age | Prevalence at Live Birth |
|---|---|
| 25 years old | 1/1350 |
| 30 years old | 1/890 |
| 35 years old | 1/355 |
| 40 years old | 1/97 |
| 45 years old | 1/23 |
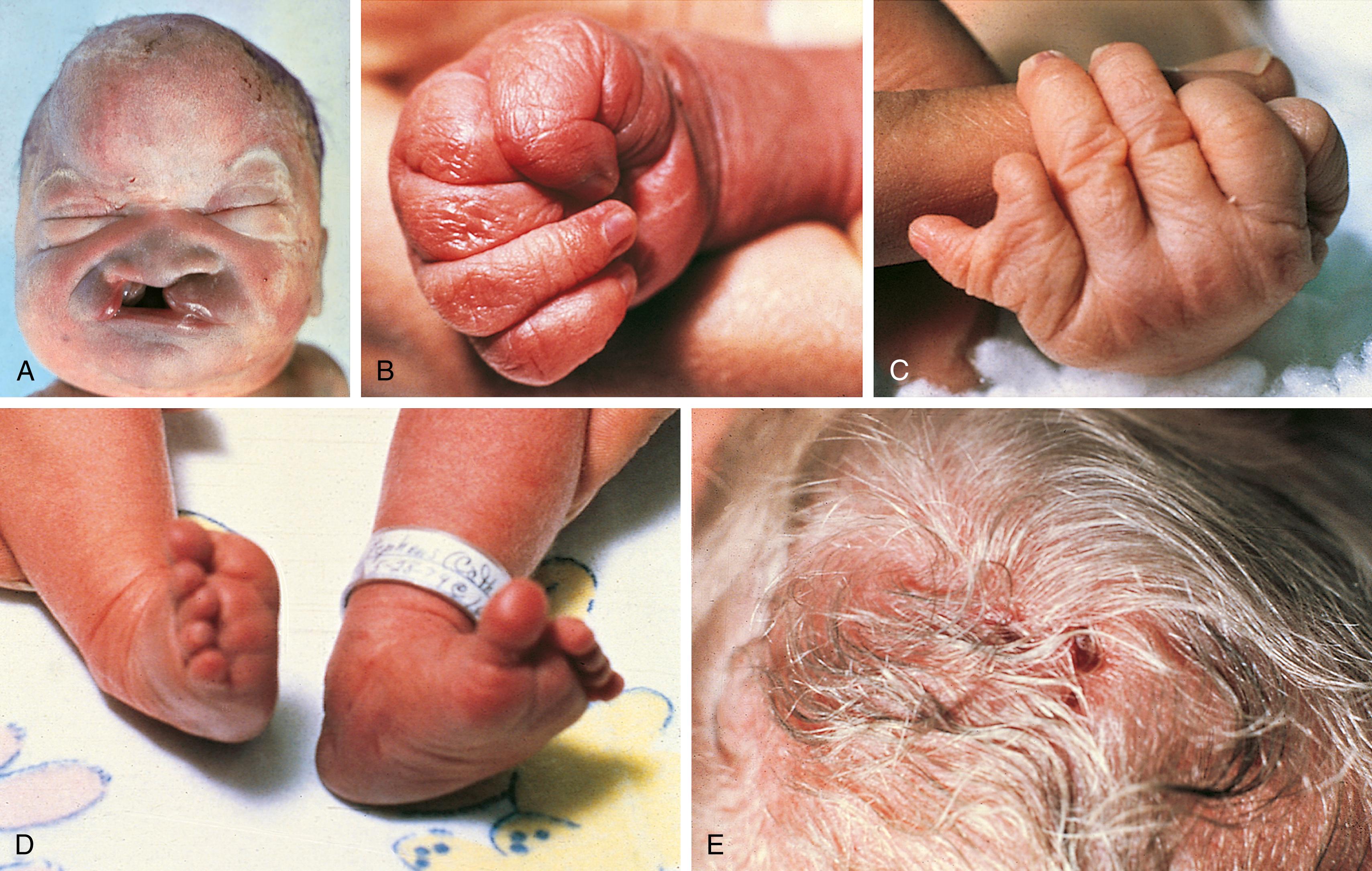
Trisomy 13 is a relatively rare (1 in 5000) genetic condition caused by the presence of additional chromosome material from all or a large part of chromosome 13. The vast majority of embryos with classic trisomy for a complete 13th chromosome abort spontaneously, but approximately 5% survive to be liveborn. Of these, about 5% survive the first 6 months of life with rare longer survival. They have a severe, recognizable pattern of malformation that allows clinicians to suspect this etiology immediately ( Fig. 1.16 , Table 1.5 ). The most recognizable features include holoprosencephaly with midline deformities, cutis aplasia, and post-axial polydactyly.
| Abnormality | Trisomy 13 | Trisomy 18 |
|---|---|---|
| Severe developmental delay/intellectual disability | †††† | †††† |
| Approximately 90% die within first year | †††† | †††† |
| Cryptorchidism in males | †††† | †††† |
| Low-set, malformed ears | †††† | †††† |
| Multiple major congenital anomalies | †††† | †††† |
| Prominent occiput | † | †††† |
| Cleft lip and/or palate | ††† | † |
| Micrognathia | †† | ††† |
| Microphthalmos | ††† | †† |
| Coloboma of iris | ††† | † |
| Short sternum | † | ††† |
| Rocker-bottom feet | †† | ††† |
| Congenital heart disease | †† | †††† |
| Scalp defects | ††† | † |
| Flexion deformities of fingers | †† | †††† |
| Polydactyly | ††† | † |
| Hypoplasia of nails | †† | ††† |
| Hypertonia in infancy | † | ††† |
| Apneic spells in infancy | ††† | † |
| Midline brain defects | ††† | † |
| Horseshoe kidneys | † | ††† |
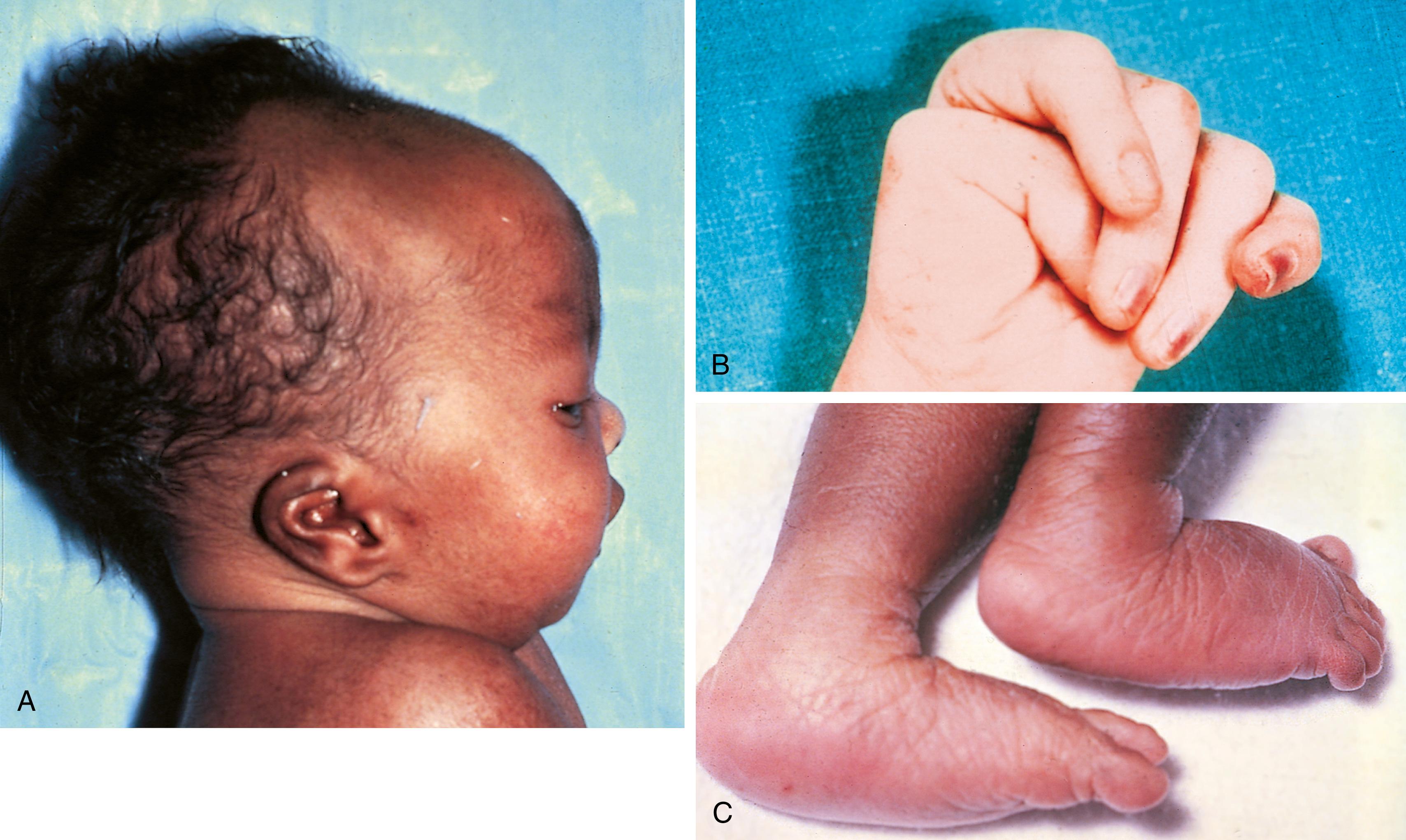
The chromosomal disorder trisomy 18 occurs in approximately 3 in 10,000 newborns, and females are more likely to be liveborn. Affected infants exhibit some hallmark features including clenched hands with overlapping fingers and are typically small for gestational age with a frail appearance and failure to thrive ( Fig. 1.17 , see Table 1.5 ). Trisomy 18 was previously thought to be almost invariably fatal in the neonatal period, however more recent data suggest that a small percentage of children can live longer; between 5% and 10% survive at least 1 year, and a few long-term survivors have been reported (frequently females with fewer structural abnormalities of major organs). Embryonal cancers such as Wilms tumor appear increased in survivors.
Partial chromosome abnormalities involving extra material determined to originate from chromosome 13 or 18 must be identified and distinguished from classic trisomies, because the clinical phenotype and prognosis may be different and, in some cases, less severe. Children with mosaicism, that is, with a normal cell line and a trisomy cell line, as well as those with trisomy of part of chromosome 13 or 18, can be identified by chromosome analysis and often have a less severe course.
Even with optimal neonatal, pediatric, and surgical management and excellent home-based care, children with classic trisomies 13 and 18 often “fail to thrive” and have significant developmental and cognitive impairments. Discussions with parents about interventions must take into account the slim possibility of long-term survival and require sensitivity to the needs of the child and family. Great care must be taken in providing a balanced picture to the family when discussing treatment options. It has been our experience that parent support organizations can be extremely helpful to family members in the long process of adjustment to having a child with a chromosome problem, or to provide a source of support through the grieving and healing process if the child dies.
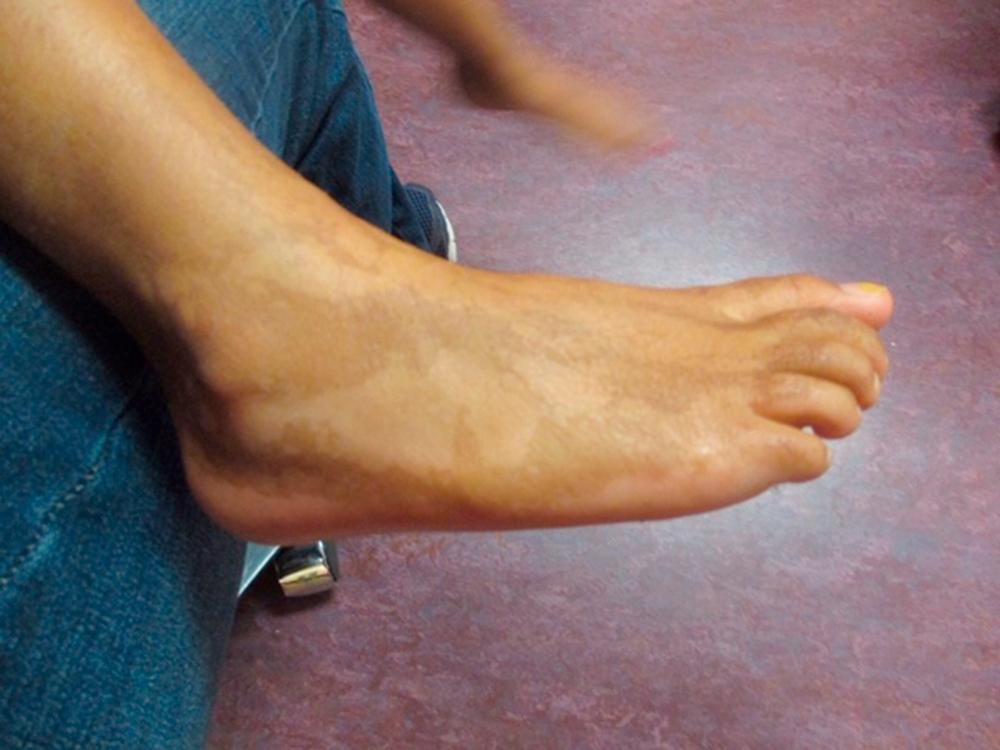
Turner syndrome is one of the three most common chromosomal abnormalities found in early spontaneous abortions. The phenotype is female. About 1 in 2000 liveborn females has Turner syndrome. Primary amenorrhea, sterility, sparse pubic and axillary hair, underdeveloped breasts, and short stature (4½ to 5 ft) are the usual manifestations. Other external physical features may include webbing of the neck; cubitus valgus; a low-set posterior hairline; a shield chest with widely spaced nipples; and malformed, often protruding, ears ( Fig. 1.18 ). Internally, renal anomalies may be present along with congenital heart disease, particularly bicuspid aortic valve (in 30% of cases) and coarctation of the aorta (in 10% of cases). Affected women have an underdeveloped uterus, and their ovaries consist only of strands of fibrous connective tissue. Newborns often have lymphedema of the feet and/or hands (see Fig. 1.18C ), which can reappear briefly during adolescence. Mental development is usually normal. Schooling and behavioral problems seem to be the same as in age-matched control subjects, although difficulty with spatial orientation (such as map reading) may be a problem. The classic physical findings of Turner syndrome may be absent, or the abnormalities may be so minimal in the newborn that the diagnosis is missed. The first indication may be unexplained short stature in later childhood or failure to develop secondary sex characteristics by late adolescence. Thus a chromosome study is indicated as part of the diagnostic workup of adolescent girls with these complaints.
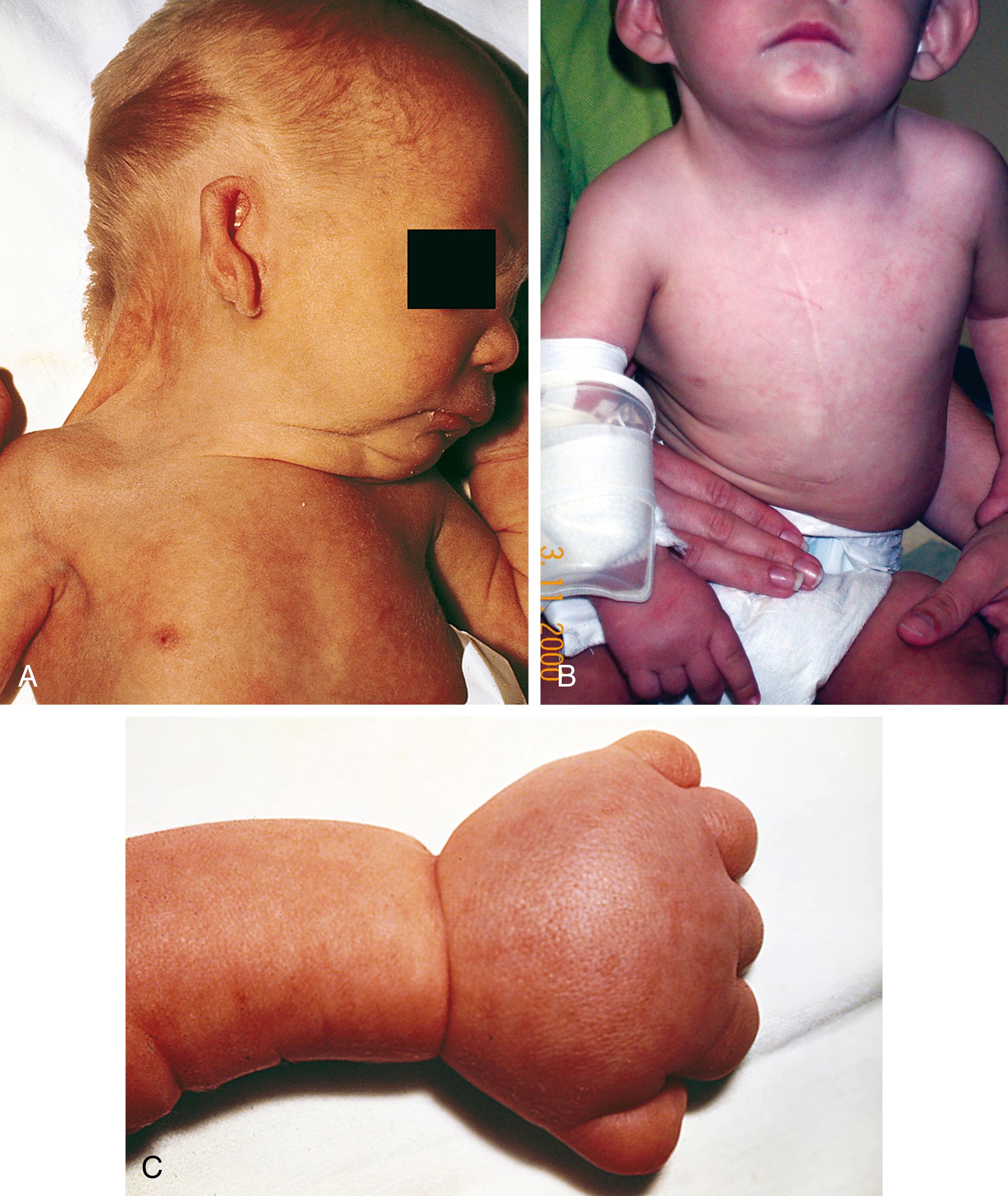
The karyotype in the majority of individuals with Turner syndrome is 45,X. Most often, the missing sex chromosome is paternally derived; the risk of Turner syndrome does not increase with maternal or paternal age. Another 15% of individuals with Turner syndrome have mosaicism (X/XX, X/XX/XXX, or X/XY). The physical stigmata may be less marked in mosaicism, in some cases with preserved fertility. The short (p) arm of the X chromosome appears most critical; deletion of the long (q) arm usually produces only streak (fibrous) gonads with consequent sterility, amenorrhea, and immature secondary sex characteristics but without the other somatic stigmata of Turner syndrome. A smaller percentage of cases of Turner syndrome have 46 chromosomes, with one normal plus one structurally abnormal X (frequently a shortened or missing short [p] arm). These cases are more likely to have other, more serious major anomalies, including cognitive deficits. A structurally abnormal X chromosome may lead to abnormal X inactivation, resulting in a deleterious dosage effect for X-linked genes. Occasionally the patient will have a 46, XY karyotype, but vital segments of the p arm of the Y chromosome have been deleted. If an XY cell line is present, the intraabdominal gonads should be removed, because they are prone to malignant change.
If the diagnosis is clinically suspected, a chromosome karyotype should be ordered. Should the affected child be 45,X or a mosaic, the parental risk for recurrence of a chromosomally abnormal liveborn is 1% to 2% but may be higher if a parent carries a structurally abnormal X chromosome. Antenatal diagnosis of chromosomally abnormal fetuses should be discussed with the parents, and the relatively good prognosis for Turner syndrome liveborns should not be overlooked. Girls with Turner syndrome should receive appropriate hormone therapy during adolescence to enable development of secondary sex characteristics and stimulate menses. Rarely, 45,X women with Turner syndrome have been fertile for a limited number of years.
One in 500 newborn boys has a 47,XXY karyotype, also known as Klinefelter syndrome. The physical stigmata are subtle and usually not obvious until puberty, at which time the normal onset of spermatogenesis is blocked by the presence of two X chromosomes. Consequently the germ cells die, the seminiferous tubules become hyalinized and scarred, and the testes become small. Testosterone levels are below normal adult male levels, although the level varies from case to case (the average being about half as much as normal). Hence there is a wide range in degree of virilization. Presentation can range from the classic tall male with a small penis and gynecomastia ( Fig. 1.19 ), to a sexually functional male with unexplained infertility. Scoliosis may develop during adolescence. The average full-scale IQ of men with Klinefelter syndrome is 98, which is about the same as the general population. Behavioral problems may be more common than in the population at large, however.
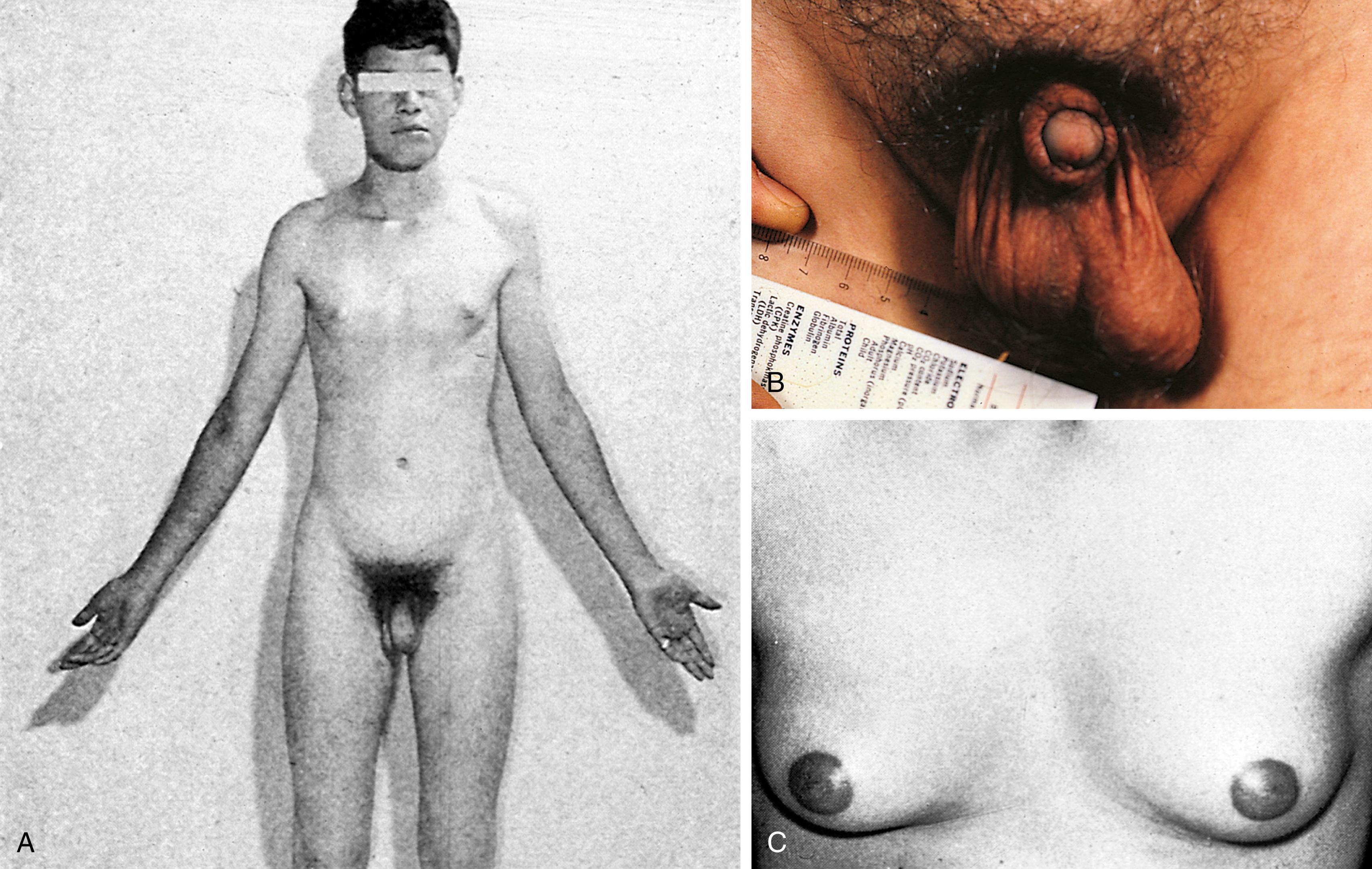
The karyotype in Klinefelter syndrome is XXY in 80% of cases and mosaic (XY/XXY) in the other 20%. Rarely the latter type may be fertile. About 60% of cases reflect a chromosome error in oogenesis, and an error in spermatogenesis occurs in 40%. The risk of having an affected child increases with maternal age. Males with more than two X chromosomes (XXXY, XXXXY) are usually cognitively impaired and are more likely to have skeletal and other major congenital anomalies, such as cleft palate, congenital heart disease (particularly a patent ductus arteriosus), and microcephaly. The parents’ recurrence risk for another chromosomally abnormal liveborn is 1% to 2%; antenatal diagnosis with subsequent pregnancies is possible.
Females with a karyotype of 47,XXX have triple X syndrome. The incidence is approximately 1 in 1000 liveborn females. Affected individuals have no characteristic abnormal physical features. Although usually within the normal range of intelligence, their IQ scores may be lower than those of their normal siblings. Delays in development of motor skills and coordination are common, and approximately 60% require some special education classes. Behavioral problems occur in approximately 30% and are usually mild. XXX women are fertile, and their children are usually chromosomally normal. The presence of additional X chromosomes (48,XXXX, or 49,XXXXX) typically results in cognitive impairment to some degree.
XYY males have a karyotype of 47,XYY. The incidence is 1 in 840 liveborn males. These males tend to be tall in comparison with their own family members, but generally their phenotypic appearance is normal. As for 47,XXX females, their IQ is usually within the normal range but may be lower than that of siblings. Affected boys often come to medical attention because of problems with fine motor coordination, speech disorders, and learning disabilities. Early reports raised concerns about significant behavioral problems; however, long-term prospective studies now suggest that these boys do not have any greater incidence of problem behaviors than the general population.
The risk of recurrence for a couple with a child with an XXX or XYY karyotype depends on many factors including the parents’ own karyotype results and advancing maternal age. Therefore it is recommended that they be referred for individualized genetic counseling when considering future pregnancies.
FISH is a laboratory technology that has revolutionized the diagnostic capabilities of clinical cytogenetic laboratories. In this technique, a DNA probe is tagged with a label that fluoresces when viewed under a special microscope. A cocktail of many repetitive DNA probes blanketing a specific chromosome from end to end can be obtained. Using special microscope filters, a clinician can simultaneously FISH paint a slide with probes fluorescing in two or three different colors. This specific FISH technique using multiple probe colors is used for specific concerns (e.g., marker chromosome identification in cancer cells). FISH and other molecular techniques are now used primarily to confirm the imbalances detected by array-based testing (see following section).
Some of the well-recognized syndromes described initially by FISH probes for chromosome microdeletion syndromes include the following: Prader-Willi/Angelman syndrome: del 15q11–13, DiGeorge sequence/velocardiofacial syndrome: del 22q11.2, Williams syndrome: del 7q11.23 and others ( Figs. 1.20–1.23 ; Table 1.6 ).
| Syndrome | Major Findings | Comments |
|---|---|---|
| Cri du chat (deletion 5p15.2) | Microcephaly, round face, down-slanting palpebral fissures, epicanthal folds, hypertelorism, catlike cry in infancy | |
| Isolated lissencephaly | Lissencephaly (incomplete development of brain with smooth surface) | Approximately 30% have deletion 17p13.3 |
| Miller-Dieker phenotype with lissencephaly | Microcephaly, lissencephaly, variable high forehead, vertical furrowing of central forehead, low-set ears, small nose with anteverted nostrils, congenital heart disease, poor feeding | Deletion 17p13.3 in vast majority |
| Deletion 22q11.2 | Phenotypes:
|
Appears to be a common deletion and should be considered in the differential diagnosis of children with multiple anomalies even if the features are not classic to any one phenotype |
| Wolf-Hirschhorn (deletion 4p16.3) | Moderate to severe cognitive impairment, hypertelorism, preauricular pit or tag, broad nasal bridge, micrognathia, cleft palate, short philtrum, growth deficiency | |
| Smith-Magenis (deletion 17p11.2) | Brachycephaly, flat facies, broad nasal bridge, short stature | Self-hugging behaviors, sleep disturbances |
In addition to classic cytogenetics, molecular cytogenetic methods are being incorporated in clinical settings at an increased rate. More recently, conventional cytogenetics is being enhanced with high-resolution molecular karyotyping using array-CGH. Array-CGH analyses are proficient in detecting imbalances in the genome and enable detection of copy number changes at high resolution. The degree of detection for copy-number variants for various cytogenetic techniques is shown in Table 1.7 . Array-CGH has been recommended by the American College of Medical Genetics (ACMG) as the first step in the investigation of patients with idiopathic intellectual disability, developmental delay, autism spectrum disorders, and multiple congenital anomalies, and has a much higher diagnostic yield—up to approximately 15% to 28%, compared to traditional G-banded karyotypes (on the order of 3%, excluding Down syndrome and other recognizable chromosomal syndromes). In addition, molecular cytogenetic techniques, such as array-CGH, have demonstrated that approximately 20% of apparently balanced chromosome translocations, de novo or familial, have gain or loss of genetic material at the breakpoints. Thus, molecular cytogenetic studies can more completely characterize the location of the chromosome breakpoints and potentially identify additional genetic material that may be duplicated or deleted that would not otherwise be detected by the traditional cytogenetic methods. By combining the array-CGH technique with classic cytogenetic and confirmatory FISH and appropriate molecular analyses, we are also able not only to identify cryptic genomic alterations but also to further analyze gross genomic alterations, including marker chromosome or other rearrangements identified by the classic cytogenetic analysis.
| Method | Resolution |
|---|---|
| Metaphase G-banded chromosome analysis (500 bands) | 5.0–10 Mb |
| Prometaphase G-banded chromosome analysis (1000 bands) | 3.0–5.0 Mb |
| Fluorescence in-situ hybridization (FISH) | 100 Kb |
Cytogenomic arrays, including:
|
As small as 10 Kb |
| Genomic Data | |
| Nuclear genome | 3.1 Gb (20,000 genes; 1 gene per ∼120 Kb) |
| Chromosome size | 48–249 Mb |
| Average gene size | 10–15 Kb (vary between 0.2 and 2500 Kb) |
| Mitochondrial genome | 16.569 Kb (37 genes) |
The application of whole-genome array-CGH in establishing the specific genetic defect has important consequences for genetic counseling of the families and follow-up of the patients. Detailed molecular analysis of the rearranged regions may help to identify the gene(s) associated with a specific phenotypic presentation. With microarray testing, many new microdeletion and microduplication syndromes have emerged (e.g., deletion 1p36, deletion 1q21.1, and deletion 16p13.11 syndromes) (see Fig. 1.22 ; and eFigs. 1.1–1.3 ). These diagnoses impact the patients’ immediate and long-term clinical management.
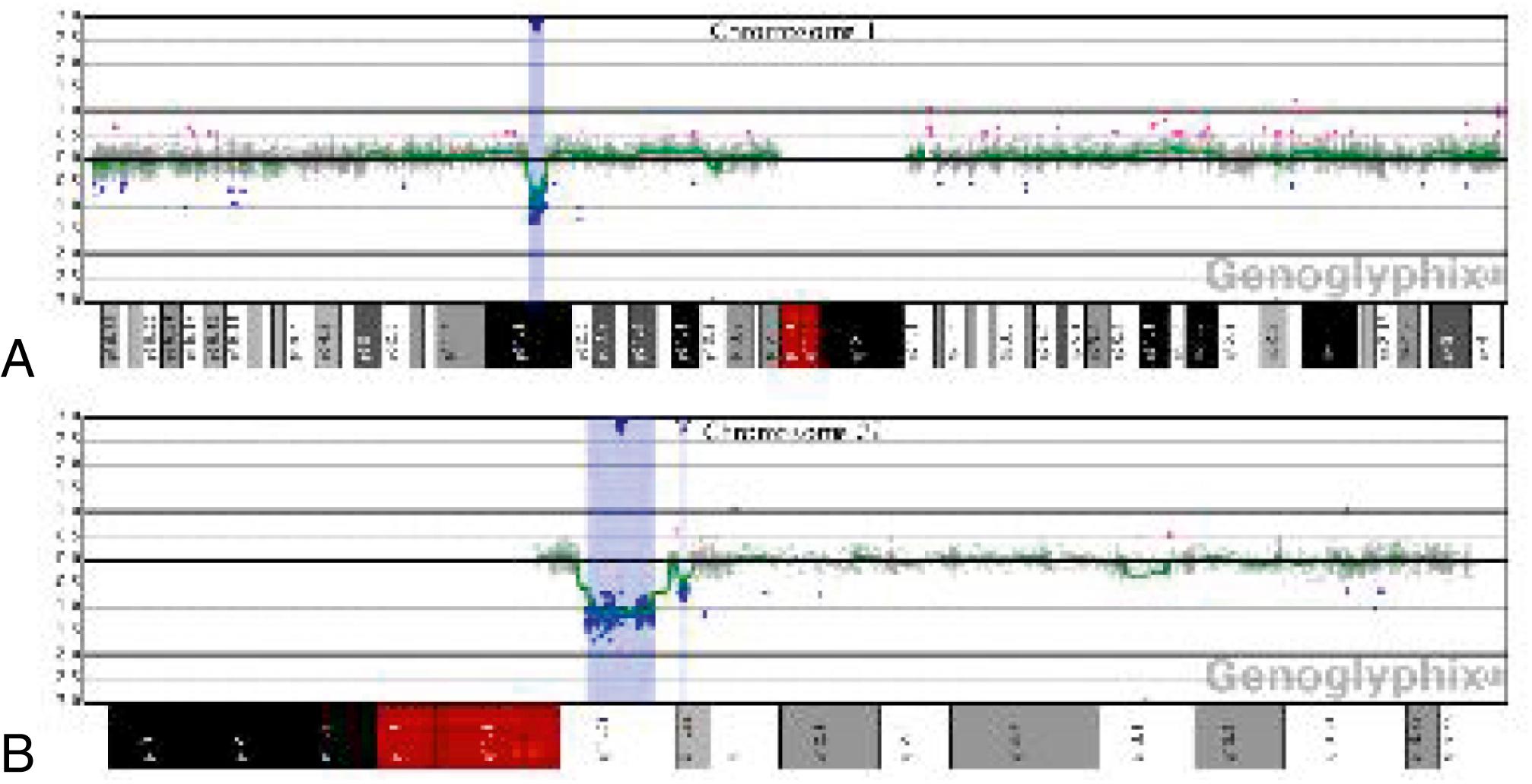
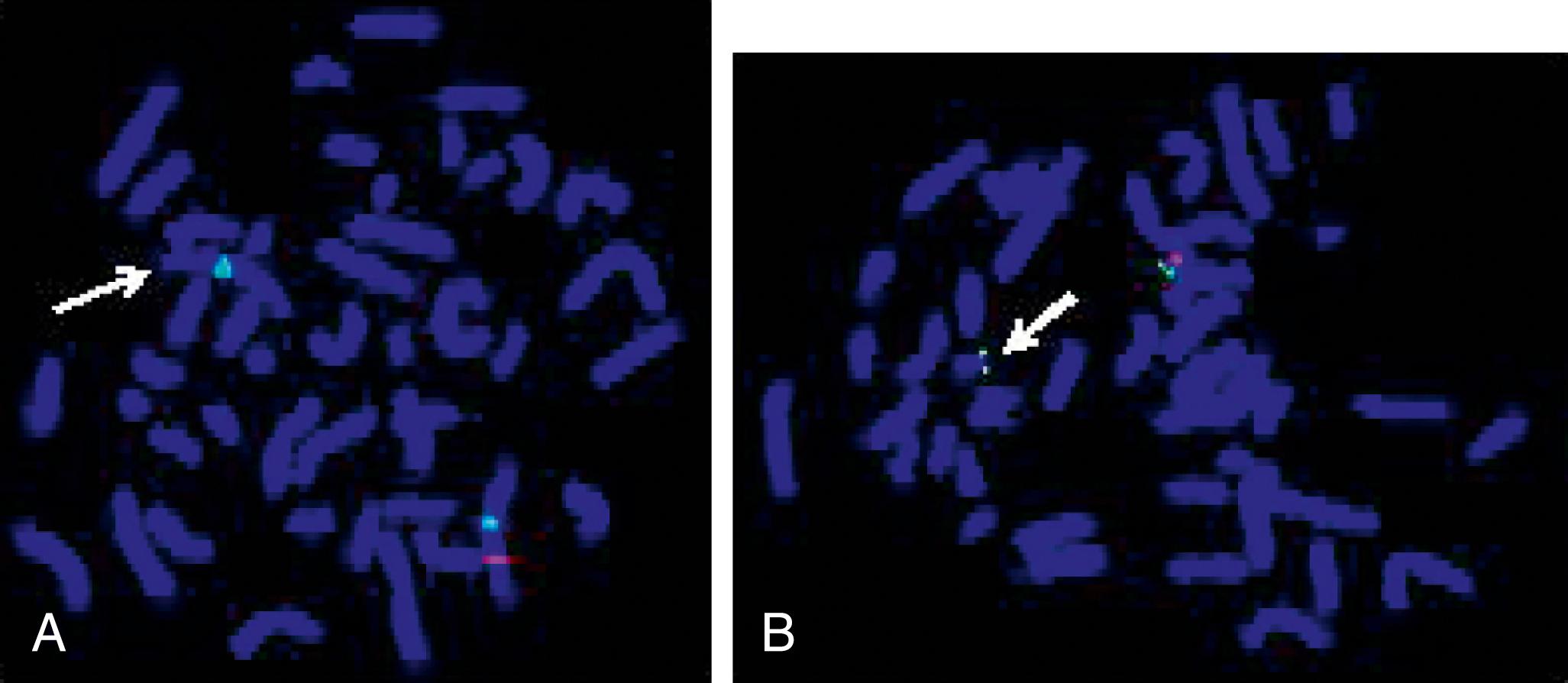
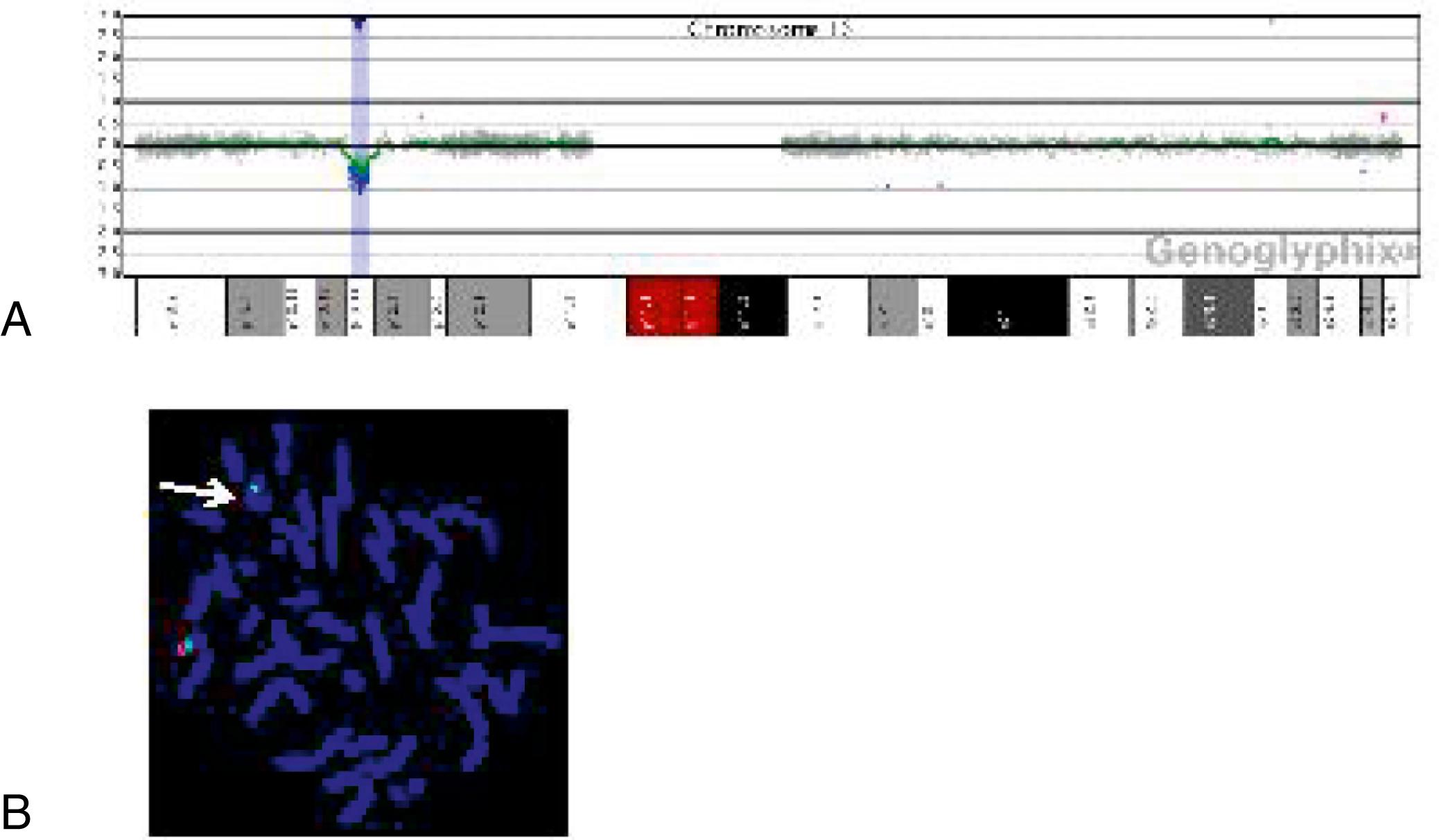
Single nucleotide polymorphism (SNP) arrays are being used in clinical settings, usually in conjunction with array CGH, and enhance genome-wide copy number analysis. The identification of “copy-number neutral” regions of homozygosity can reveal consanguinity and suggest genes of interest in the homozygous regions. This molecular cytogenetics test therefore simultaneously interrogates and not only identifies copy number variations (CNVs) but is also capable of detecting region(s) with long or short stretches of homozygosity (“regions of homozygosity” [ROH]) known as DNA copy number neutral alterations. Genetic counseling is helpful in interpretation, particularly for variants of uncertain significance or in the unanticipated identification of non-paternity or possible consanguinity.
The impact of these newer methodologies continues to emerge, and their usefulness in providing information key to clinical prognosis is clearly becoming evident. Molecular karyotyping and SNP arrays are ultimately more cost-effective tests and have been extremely useful to clinicians in identifying necessary medical surveillance and treatment options, and they provide information on recurrence risks and prenatal options for families. Recommendations include but are not limited to specific surveillance, pharmacological treatment, cancer-related screening or exclusion of screening, contraindications, and referrals for further evaluation.
Molecular cytogenetic techniques have identified a group of disorders typically resulting in a well-defined and recognizable presentation, due to submicroscopic deletions or duplications involving a number of sequential genes. A number of these may be commonly recognizable in the clinical setting, such as DiGeorge syndrome (see also Chapter 4 ), Williams syndrome (see also Chapter 5 ), and Angelman and Prader-Willi syndromes (covered later in this chapter). The remaining syndromes are outlined briefly in Table 1.6 and in Figs. 1.20–1.22 .
It has long been recognized that there is a significant excess of males having intellectual disability. Much of this inordinate male representation is the result of altered X-linked recessive genes. These may represent new mutations or inheritance of the abnormal gene from normal heterozygous (carrier) mothers. About 1 in 150 individuals, usually male, has some form of X-linked intellectual disability. Of these, it is estimated that between 30% and 50% have fragile X syndrome, translating to an incidence of 1 in 4000 males.
In 1969, Herbert Lubs noted in low folate lymphocyte cultures an apparent break (a “fragile site”) on the long arm of the X chromosome (Xq27) (see Fig. 1.23 ). Fragile X syndrome is the first recognized example of a trinucleotide repeat disorder, with the clinical significance of the “fragile site” confirmed in 1977. The gene involved, located at Xq27.3, is called FMR1 and is active in brain cells and sperm. At the start of the gene is a DNA trinucleotide cytosine-guanine-guanine (CGG) motif, which in the general population is normally repeated about 5 to 50 times (the average being 30). When there are more than about 55 repeats, the region can become unstable during oogenesis such that the number of repeats increases during oogenesis. In the presence of 55 to 200 CGG repeats, the individual has a fragile X premutation . Individuals with a premutation appear clinically and cognitively normal in early life but can develop ataxia in later adult life (fragile X ataxia syndrome, aka FRAXTAS). When the premutation expands to greater than 200 repeats, gene expression is impaired, and the individual has a full mutation, resulting in fragile X syndrome in males and a rare subset of females ( Fig. 1.24 ).
No cases of new mutations for these FMR1 gene CGG trinucleotide expansions or repeats have been found. That is, all such expansions are inherited from a parent, but it appears expansion of the number of repeats occurs during oogenesis, but not spermatogenesis. The daughters of men with premutations are obligate premutation carriers and are at risk to have sons with full mutations. The relative risk for a woman to pass down a pre- or full mutation is shown in Table 1.8 .
| Number of CGG Repeats in Mother’s Premutation | Risk of Expansion to Full Mutation in Offspring (%) |
|---|---|
| 55–59 | 3.7 |
| 60–69 | 5.3 |
| 70–79 | 31.1 |
| 80–89 | 57.8 |
| 90–99 | 80.1 |
| 100–109 | 100 |
| 110–119 | 98.1 |
| 120–129 | 97.2 |
Males affected with fragile X syndrome have cognitive impairment, ranging from severe to borderline in degree. The majority have an IQ between 20 and 49, and the remainder fall in the 50 to 70 IQ range. Furthermore, IQ may decline with age. The majority have speech delay, short attention span, hyperactivity, persistence of mouthing objects, and poor motor coordination. Many exhibit a variety of disordered behaviors, including disciplinary problems, temper tantrums, poor eye contact, perseverative speech, hand flapping, avoidance of socialization, and rocking. Physical stigmata may include long, wide, or protruding ears; long face; a prominent jaw; flattened nasal bridge; “velvety” skin; hyperextensible joints; and mitral valve prolapse. Relative macrocephaly is more likely than microcephaly. Macroorchidism is found in most mature males.
Approximately 50% of females affected with full-mutation fragile X are clinically normal. The 50% who are affected usually have lesser degrees of cognitive impairment than males; about 35% fall in the 20 to 49 IQ range, and the remainder falls in the 50 to 70 mild intellectual disability range. However, learning disabilities, mood disorders, schizoid personality, and significant disturbances in affect, socialization, and communication are common. The physical features often seen in males with fragile X syndrome are less common in females.
Laboratory testing for fragile X mutations is done by molecular genetic techniques, by Southern blot or polymerase chain reaction (PCR) analysis of DNA. These techniques can also be applied to fetal cells for the purpose of antenatal diagnosis. However, if the fetus is a female with a full mutation, it is impossible to predict whether the child will be clinically affected with fragile X syndrome because of random X inactivation.
Rarely, an individual may seem to have a mild form of fragile X syndrome, but tests are negative using these molecular genetic laboratory techniques. Another fragile X gene site (FRAXE) distal to the fragile X gene on Xq is associated with mild intellectual disability and a positive fragile X testing.
The number of known trinucleotide expansion disorders is increasing ( Table 1.9 ). Three other examples are Huntington disease, caused by an intragenic CAG trinucleotide expansion; myotonic dystrophy, type 1, resulting from a 3′ untranslated region CTG expansion; and Friedreich’s ataxia caused by an intronic GAA expansion.
| Condition | Nucleotide Repeat Expansion | Inheritance | Gene | Symptoms |
|---|---|---|---|---|
| Huntington disease | (CAG)n | AD | HTT | Choreoathetosis, dementia |
| Kennedy disease (Bulbospinal muscular atrophy) | (CAG)n | X-linked | AR | Muscle weakness, cramps, fasciculations of face, difficulty swallowing |
| Dentatorubral-pallidoluysian atrophy | (CAG)n | AD | ATN1 | Myoclonic epilepsy, choreoathetosis, dementia, brain degeneration |
| Machado-Joseph disease (Spinocerebellar ataxia, type 3) | (CAG)n | AD | ATXN3 | Progressive cerebellar ataxia, peripheral neuropathy, dysphagia, ophthalmoplegia, Parkinsonian-like features |
| Spinocerebellar ataxia, type 1 | (CAG)n | AD | ATXN1 | Progressive cerebellar ataxia, peripheral neuropathy, dysphagia, ophthalmoplegia |
| Spinocerebellar ataxia, type 2 | (CAG)n | AD | ATXN2 | Progressive cerebellar ataxia, dysphagia, retinal disease, ophthalmoplegia, peripheral neuropathy |
| Spinocerebellar ataxia, type 6 | (CAG)n | AD | CACNA1A | Progressive cerebellar ataxia, dysphagia, hemiplegic migraine |
| Spinocerebellar ataxia, type 7 | (CAG)n | AD | ATXN7 | Progressive cerebellar ataxia, dysphagia, retinal disease, ophthalmoplegia |
| Spinocerebellar ataxia, type 17 | (CAG)n | AD | TBP | Progressive cerebellar ataxia, dysphagia, nystagmus, ophthalmoplegia, psychiatric manifestations |
| Myotonic dystrophy, type 1 | (CTG)n | AD | DMPK | Weakness, myotonia, cataracts, hair loss, cardiac arrhythmia, infantile hypotonia, respiratory insufficiency, ID |
| Spinocerebellar ataxia, type 8 | (CTG)n | AD | ATXN8 | Progressive cerebellar ataxia, nystagmus |
| Fuchs corneal dystrophy, type 3 | (CTG)n | AD | TCF4 | Progressive corneal opacification, vision loss |
| Friedreich ataxia | (GAA)n | AR | FXN | Progressive ataxia, dysphagia, cardiac arrhythmias |
| Oculopharyngeal muscular dystrophy | (GCG)n | AD | PABPN1 | Progressive dysphagia and coughing, ptosis |
| Myotonic dystrophy, type 2 | (CCTG)n | AD | ZNF9 | Weakness, myotonia, cataracts, hair loss, cardiac arrhythmia, infantile hypotonia, respiratory insufficiency, ID, endocrinopathies |
| Spinocerebellar ataxia, type 10 | (ATTCT)n | AD | ATXN10 | Slowly progressive cerebellar ataxia, seizures |
| Spinocerebellar ataxia, type 31 | (TGGAA)n | AD | BEAN | Progressive cerebellar ataxia, nystagmus, hearing loss |
| Spinocerebellar ataxia, type 36 | (GGCCTG)n | AD | NOP56 | Progressive cerebellar ataxia, nystagmus, hearing loss, tongue fasciculations |
| Frontotemporal dementia, amyotrophic lateral sclerosis | (GGGGCC)n | AD | C9Orf72 | Dementia with executive dysfunction and abnormal behaviors, weakness |
| Progressive myoclonic epilepsy, 1A | (CCCCGCCCCGCG)n | AR | EPM1 | Ataxia, dysarthria, seizures (myoclonus, GTC, absence) |
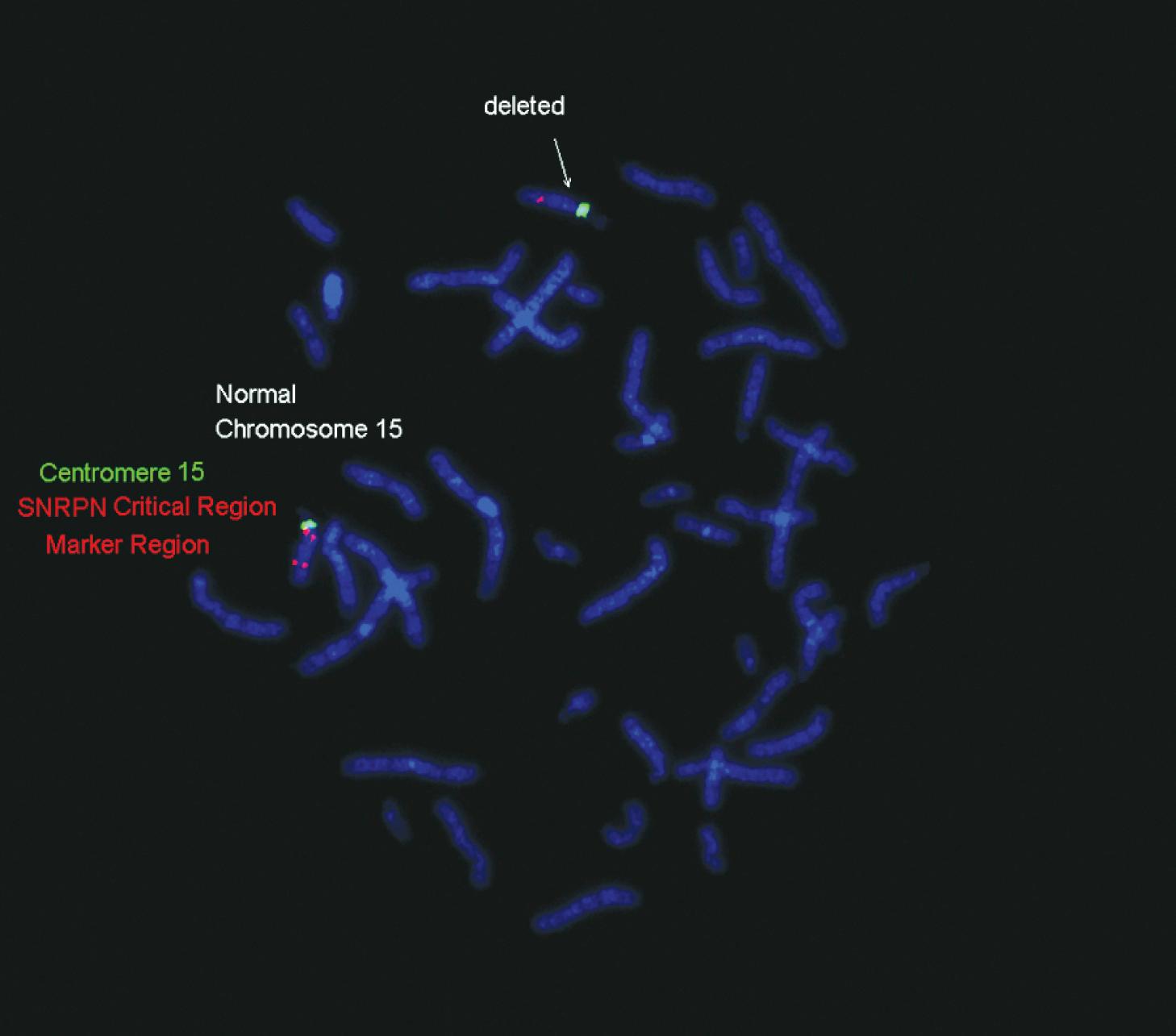
The concept of imprinting refers to the fact that the function of certain genes is dependent on their parental origin: maternal versus paternal. One prominent example is the 15q11–q13 region of chromosome 15, a region that contains several imprinted genes that, when abnormal, result in recognizable constellations of physical and behavioral problems. Prader-Willi and Angelman syndromes are disorders that derive from abnormalities of these imprinted genes. Absence of the paternally derived region of this chromosome results in Prader-Willi syndrome, while absence of the maternally derived region of this chromosome results in Angelman syndrome.
Mechanisms that can produce the Prader-Willi phenotype include the following:
A chromosome deletion of 15q11–q13, including the Prader-Willi critical region of the paternally derived chromosome 15 (majority of cases)
A structural chromosome abnormality involving the Prader-Willi critical region of 15q11–q13 (translocation, etc.)
Maternal UPD in which the child has two maternally derived chromosome 15s and no paternally contributed chromosome 15 (25% of cases)
Mutations of imprinting control center genes (1% of cases)
The critical region of chromosome 15 for Angelman syndrome is located adjacent to the Prader-Willi critical region. However, it is the maternally derived chromosome segment that is absent. The six currently identified etiologic mechanisms of Angelman syndrome include the following:
A large chromosome deletion of 15q11–q13 including the Angelman critical region of the maternally derived chromosome 15 (68% of cases)
A structural chromosome abnormality involving the Angelman critical region of 15q11–q13 (translocation, etc.)
Paternal UPD of chromosome 15 (7% of cases)
Mutations of imprinting control center genes (3% of cases)
Mutations of the ubiquitin-protein ligase gene (UBE3A) (11% of cases)
Classic phenotype, with no identifiable etiologic mechanisms but a positive family history of other affected individuals (11% of cases)
Because of etiologic variability and complexity of the diagnostic process, families of children suspected of having either of these disorders should be referred for genetic evaluation and diagnostic testing to ensure the most accurate determination of etiologic mechanism, and therefore, of recurrence risk.
Current diagnostic testing for these disorders includes the following:
Methylation studies, which determine whether genes within the 15q11–q13 critical region are correctly marked (methylated) by their parent of origin (detects approximately 99% of PW and 80% of Angelman syndrome cases)
Chromosome microarray or FISH probes of the chromosome region (detects approximately 65% to 75% of PW and 68% of Angelman syndrome cases)
SNP array (detects 80% to 90% of Prader-Willi syndrome (PWS) and 68% to 78% of Angelman syndrome cases)
In some cases of Angelman syndrome, sequence analysis of the UBE3A gene (11%)
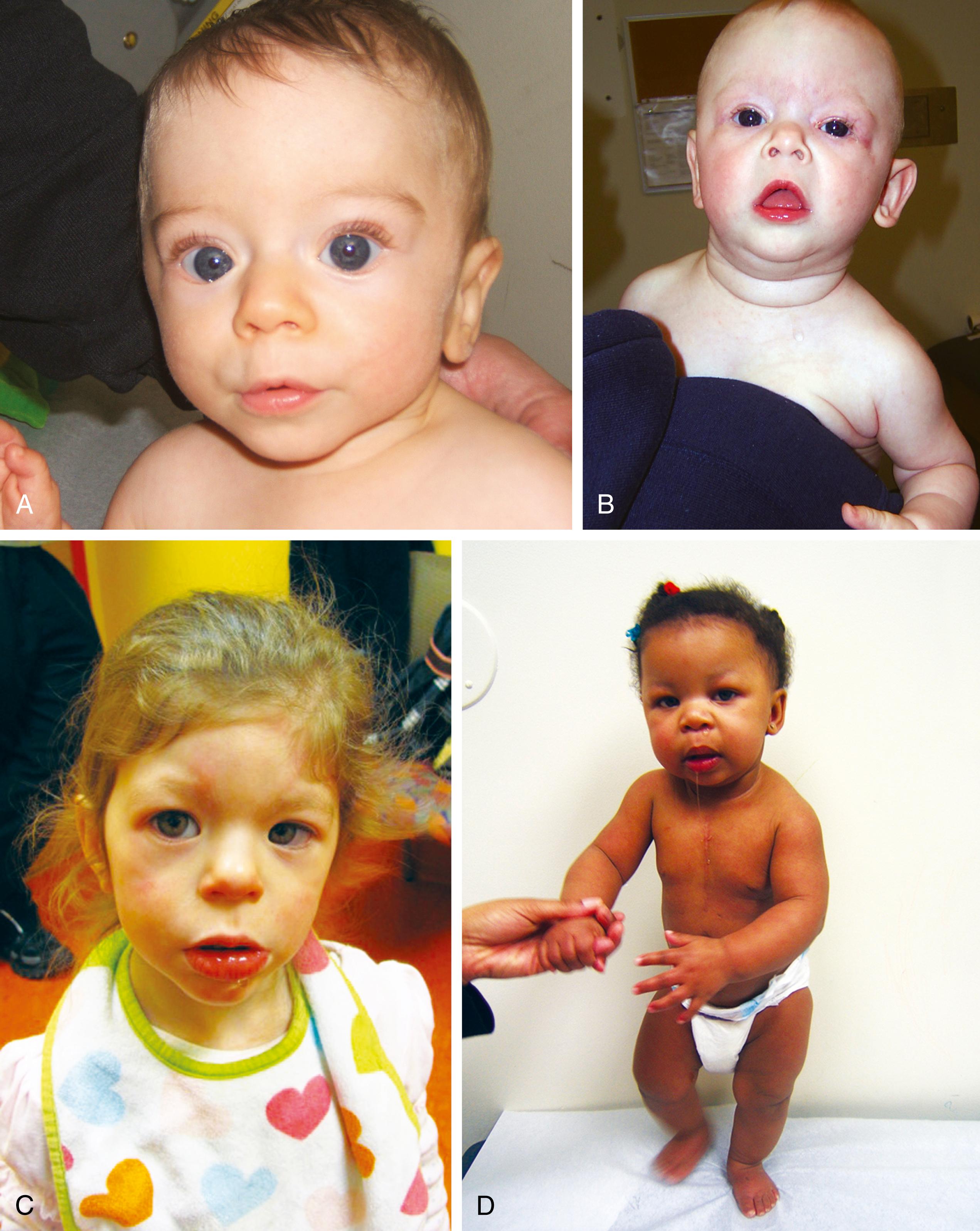
Note: Some other examples of imprinting disorders include Beckwith-Wiedemann syndrome ( Fig. 1.25 ) and Russell-Silver syndrome.
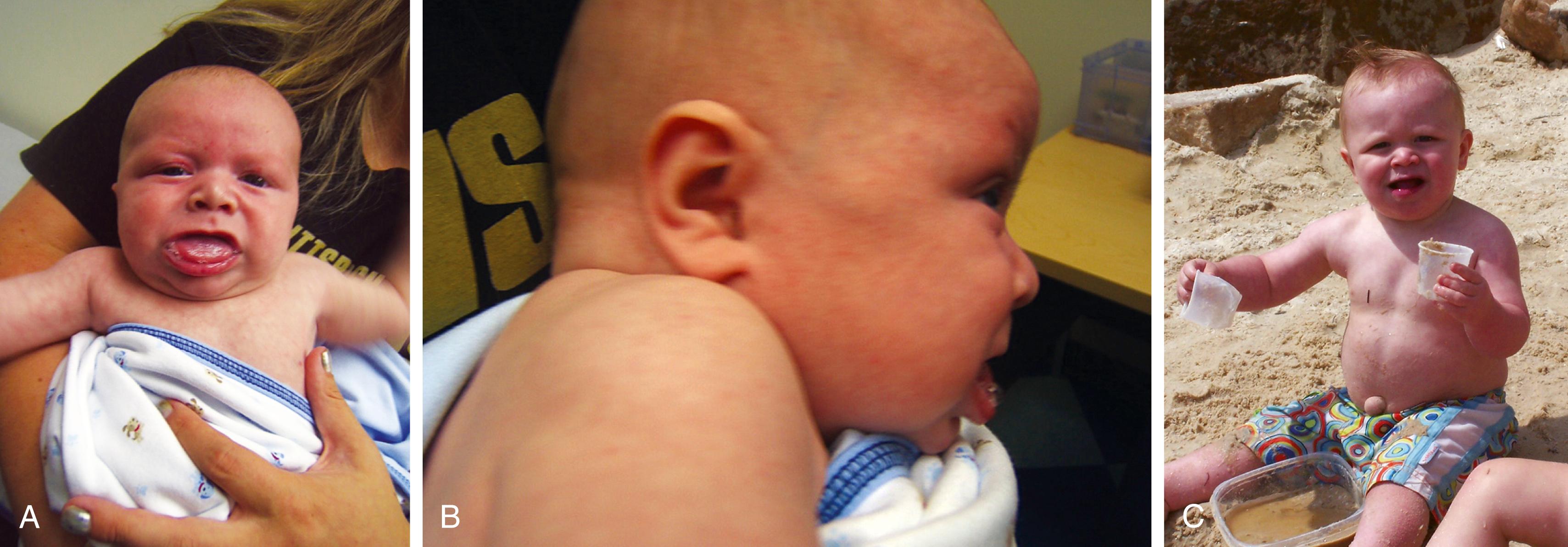
Newborns affected with Prader-Willi syndrome usually are markedly hypotonic with a weak cry, poor sucking and swallowing, decreased deep tendon reflexes, and often have a history of decreased fetal movement in utero and breech fetal position and are small for gestational age. Hypotonia can predispose to choking episodes that can cause respiratory problems. Failure to thrive is virtually ubiquitous. Subsequently motor development is delayed, speech even more so, and most patients have cognitive impairment in the mild to moderate range. Hypotonia abates over the first 2 to 3 years, and patients develop an insatiable appetite that rapidly results in morbid obesity ( Fig. 1.26 ). The distribution of excess fat is particularly prominent over the lower trunk, buttocks, and proximal limb. Subtle facial features can include narrow bifrontal diameter, “almond shaped” eyes, and strabismus. Other features include skin picking, small hands and feet, short stature, hypogenitalism, and hypogonadism. In patients with chromosome deletions, hypopigmentation is common, with blond to light brown hair, blue eyes, and sun-sensitive fair skin.
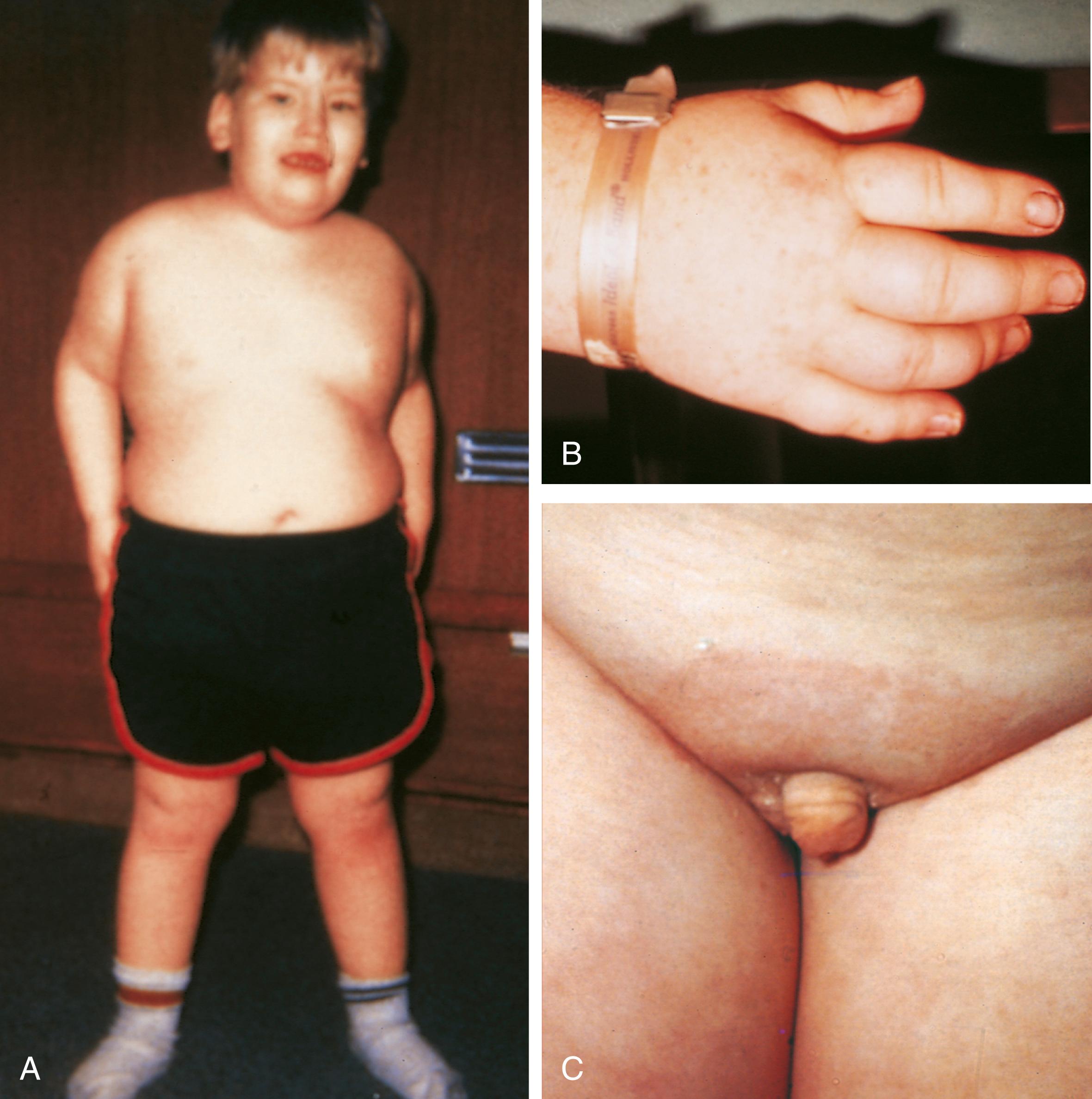
Older children with Prader-Willi syndrome can exhibit extreme emotional lability and temper tantrums. These conditions and the overeating often can be partly ameliorated by intensive inpatient behavioral modification programs followed by longitudinal parental support and follow-up in the home. Growth hormone therapy can exacerbate sleep apnea, but significantly improves weight control, and reduces associated morbidities such as diabetes or risk of Pickwickian syndrome. Interestingly, despite a normal basal metabolic rate, weight reduction requires significantly more severe caloric restriction than usual. In untreated individuals, cardiorespiratory complications from morbid obesity can reduce life expectancy, therefore early diagnosis and treatment are crucial.
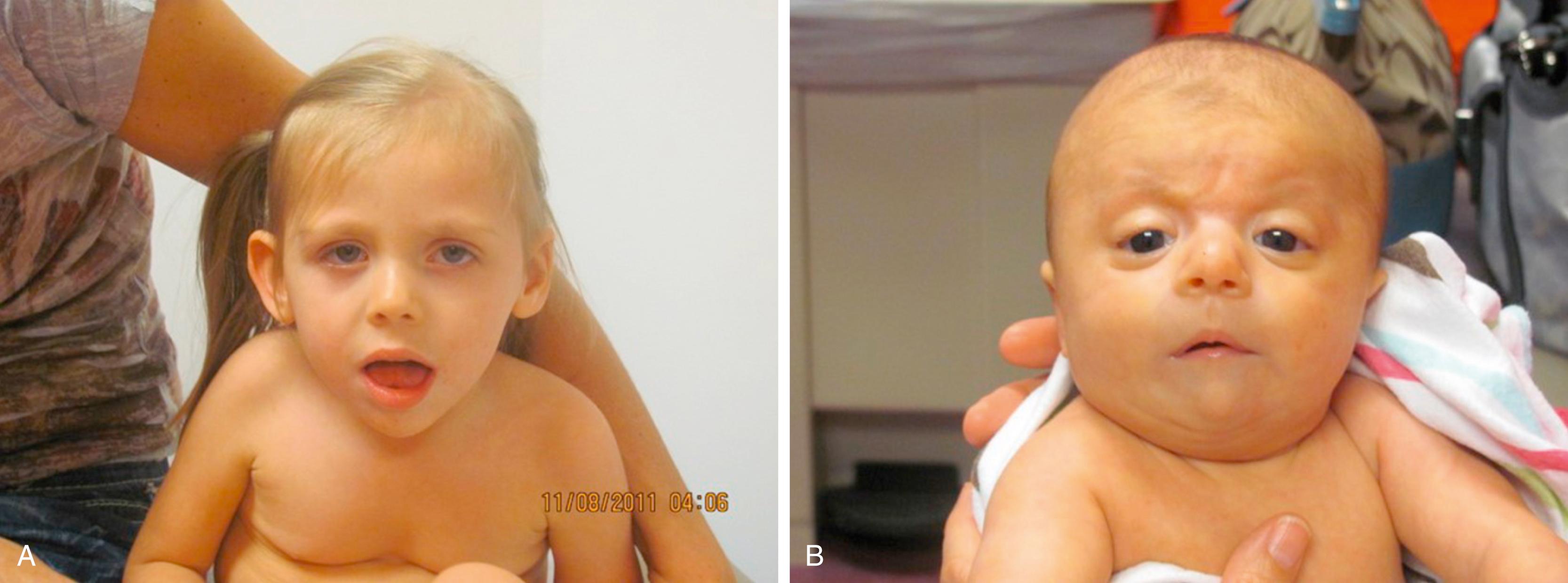
Angelman syndrome, first recognized in 1956, has an incidence of 1 in 15,000 to 1 in 20,000 live births. Except for the shared tendency to have hypopigmentation, the clinical phenotypes of Prader-Willi and Angelman syndromes are quite different. The latter have severe cognitive deficits, speech is impaired or absent, and inappropriate paroxysms of laughter are common. Physical features include microbrachycephaly, maxillary hypoplasia, large mouth, prognathism, and short stature (in adults). The gait is ataxic, with toe-walking and jerky arm movements. Akinetic or major motor seizures are common and can begin in the neonatal period. Although survival to adulthood is possible, to date only one patient with Angelman syndrome has been known to reproduce.
A gene consists of a sequence of DNA that contains the code for production of a “functional product” along with sequences that ensure proper expression of the gene. The gene product might be an RNA molecule, a polypeptide chain, or a protein that ultimately becomes a structural component of a cell or tissue, or enzyme. The latter may catalyze a step in formation or modification of another product, a step in cell metabolism, or one of a number of steps involved in the breakdown or degradation of molecules that are no longer necessary. Proper gene expression includes production of the product at the right time (e.g., during embryogenesis vs. postnatal expression), in the needed amount, in the correct cell type, and ensures its transport to its proper site of biologic action.
Approximately 20,000 genes are arranged in linear fashion on the chromosomes, all having their own specific locus (location). Genes range in length from about a thousand bases to hundreds of thousands of bases (any one of which can be subject to mutation). Genes are composed of alternating sequences of coding DNA, termed exons, interspersed with noncoding sequences of DNA, termed introns. Exons are further subdivided into triplets of bases, termed codons, each of which encodes a specific amino acid within the polypeptide product. Because there are 64 possible triplet combinations of the four nucleotide bases (adenine, guanidine, thymine, and cytosine) and 20 amino acids, most amino acids have more than one codon that can specify them, the exceptions being methionine and tryptophan, which have only one specific codon each. In addition, three triplets encode stop codons in the messenger ribonucleic acid (mRNA) that signal the termination of mRNA translation.
The process of going from DNA code to polypeptide product has many steps and begins with transcription, during which the DNA of the gene serves as a template for the formation of a mRNA molecule. This process requires RNA synthetase, proteins called transcription factors, and regulatory elements, as well as DNA sequences that signal where to start and stop transcription. After this, both ends of the mRNA molecule undergo modification where the introns are excised and the exons spliced together. Then the mRNA is transported to the rough endoplasmic reticulum within the cytoplasm, where it attaches to ribosomes, and the process of translation from mRNA template to polypeptide chain begins. During translation, transfer ribonucleic acid (tRNA) molecules, each of which is specifically designed to attach to a particular amino acid, find their target moieties and bring them into position at the correct time over a codon on the mRNA that specifies for their particular amino acid.
After assembly, the polypeptide chain is released from its template and then may be subject to posttranslational modification. Steps may include folding, bonding into a three-dimensional conformation, being combined with another or other polypeptide chains as part of a protein complex, being split into smaller segments, and addition of phosphate or carbohydrate moieties. Thereafter, it is transported to its site of action via directional terminal sequences, which are then cleaved from the finished product. Mutation of a gene encoding the polypeptide product or for any molecule used at any step along the entire process can adversely influence the end product.
A single-gene variant produces a permanent change in a gene’s DNA sequence and may involve anywhere from one to several thousands of nucleotides. The simplest mutation is a change of a single base pair, but more complex mutations also occur including change of more than one base pair, or deletion or insertion of base pairs. Examples of mutations include missense variants, in which a base substitution changes a codon specific for one amino acid into one specifying another; frameshift variants, in which a deletion or insertion is not an exact multiple of three bases, and thereby shifts the reading frame for transcription (and later translation) from that point on; and nonsense mutations, in which a base substitution changes a codon for an amino acid into one specifying one of the three possible stop codons in mRNA, thereby stopping translation prematurely. In some cases, a mutation in an intron can alter the splicing and removal of the intron, also resulting in an aberrant mRNA molecule.
Some variants have no effect on phenotype or cell function. These are referred to as benign variants . One example is a base substitution within a codon for an amino acid that changes it to another codon specifying the same amino acid. Still other variations have no adverse effect but rather encode normal variations in human characteristics (e.g., eye or hair color). Other variants do have adverse effects and are causative in disease. Though commonly referred to as a mutation, the clinically precise term is a pathogenic variant for those changes in the genetic code that result in medically significant findings. Genetic sequencing can reveal novel genetic changes of uncertain pathology, termed “variants of uncertain significance.” Computer programs attempt to predict the potential effect on the final protein, called “in silico” analyses, and the ACMG works with laboratories to provide updated variant classification. However, in some cases the effect of a DNA variant can remain uncertain until sufficient occurrences have been recorded.
Next generation sequencing (NGS) methodologies are rapidly becoming a mainstay of clinical diagnosis for heritable disorders. The first clinical application of NGS was for disease gene discovery in cancer involving analyses of patients’ tissue samples with suspected inherited monogenic aberrations in the nuclear and mitochondrial genome. Newer applications of NGS with immense promise for clinical diagnostic utility include whole genome sequencing (WGS) and diagnostic whole exome sequencing (WES). WES allows the unbiased simultaneous interrogation of the coding regions, or exons, and the exon boundaries of approximately 20,000 genes by massive parallel sequencing and is thus a powerful tool for making a diagnosis in complex clinical cases with a suspected heritable etiology that is becoming increasingly applicable to the clinical setting. Most disease-causing changes in the DNA sequence are found in the exons (exome) consisting of about 3% of the genome. WES must be interpreted in the context of familial segregation of any changes detected in the patient’s sequence, the patient’s medical and family history, and detailed knowledge of clinical features including other lab testing. WES has technological limitations in that it does not detect certain types of deleterious genetic changes, such as deletions, duplications, trinucleotide repeats, or methylation defects, and can lack sensitivity near highly repetitive regions, such as the first exon within a gene. Although mapping of the genes is complete, our understanding of the function of most genes is in its infancy. Ultimately, WES provides profound diagnostic and economic advantages in the diagnosis of pediatric and adult heritable disease presenting in the medical genetics clinic. The impact of achieving a definitive diagnosis and identification of the heritable etiology helps to streamline the multidisciplinary medical needs and implication(s) on the patient and his or her family. Diagnosis in patients with a comorbid diagnosis and blended phenotypes may only be possible through utilization of WES. Similarly, WGS and WES are critical in the identification of new mutations and genetic causes of novel phenotypic presentations ( Fig. 1.27 ).
Molecular sequencing can identify many genetic disorders, particularly if the suspected gene or disorder in question is known. Exome sequencing is increasingly utilized for the diagnosis of genetic disorders when the specific underlying genetic defect is not clearly identifiable. This technique sequences the exons (coding regions) of approximately 20,000 genes at once, with an estimated diagnostic rate of 40%. Some clinical laboratories have initiated the combined technology for exome sequencing with chromosomal array, or to replace chromosomal array as the first-line test for individuals with intellectual disability, autism, or congenital anomalies. Expanded sequencing techniques including WGS may be more efficient at detecting copy number variants simultaneously with detection of genetic mutations in highly repetitive parts of the genome, in early exons within a gene, and in deeply intronic regions (e.g., that can affect splicing, etc.), but is still emerging from the research phase to bedside genetics.
A mutation (pathogenic variant) in a gene typically results in either alteration of the amount of gene product produced, failure to produce the product at all, and/or compromise of its functional integrity. The loss of gene product or function is also referred to as haploinsufficiency . The greater the degree of functional loss, the more severe the clinical manifestations of the disorder and often the earlier their onset. A single-gene disorder is the result of a mutation altering the DNA sequence within a single gene on a chromosome. If mutation in the gene on only one chromosome is sufficient to cause disease, the disorder is dominant . If mutation of the gene on both chromosomes is required to manifest disease, the disorder is recessive . Genes on the X chromosome are called X-linked , and expression can depend on how many active X chromosomes express the gene. Figs. 1.27 through 1.46 are representative of single-gene disorders.

Pathogenic variant(s) of gene(s) within the nuclear genome are also recognized as mendelian disorders. The occurrence and/or recurrence are in fixed proportions (Mendel’s laws). These disorders are compiled in a catalog, the Online Mendelian Inheritance in Man (OMIM; http://www.ncbi.nlm.nih.gov/omim ), which is an excellent resource. Phenotype/genotype correlations are unfolded by detailed clinical evaluation, recognition at a clinical level, and confirmation by molecular diagnostics confirming the genotype.
Pedigree analyses are usually helpful in recognizing autosomal, X-linked, recessive, and dominant patterns. Gene penetrance, disease expressivity, genetic (locus) heterogeneity, and allelic heterogeneity are some of the well-recognized complexities characterizing mendelian disorders.
When the gene product is an enzyme or a component of an enzyme, this results in interruption of its step in a chain of reactions that may be involved in the formation or modification of a product, a step in cell metabolism, or in the degradation of molecules no longer needed by the cell. The missed step results in a build-up of substrate from the step preceding the one in which the affected enzyme acts. In some instances, this accumulated substrate can be toxic, as in PKU. In others, ever-expanding storage of substrate can adversely affect cell function, as in the lysosomal storage diseases. Since many enzyme disorders result in inborn errors of metabolism, these will be further considered in the section on Inborn Errors of Metabolism.
The family of disorders known as osteogenesis imperfecta (OI; see also Chapter 22 and see Fig. 1.37 ) provides a good example of the effects of pathogenic variants that alter the precursors of a structural protein, in this case type I collagen. Type I collagen is a triple helix made up of two pro-α1 chains and one pro-α2 chain. The procollagen strands are composed of hundreds of amino acid triplet repeats, with glycine (the smallest amino acid) being the first member of each triplet and forming the apex of each bend in the helical structure. A base substitution in a codon specifying glycine at any one of the hundreds of such points along either the COL1A1 gene or the COL1A2 gene can produce inadequate or unstable collagen production in one of several ways. In some cases, genetic variation results in the production of an unstable mRNA molecule that is degraded in the nucleus, or structurally abnormal collagen chains subject to degradation, resulting in inadequate collagen production that is normal in structure but reduced by 50% (haploinsufficiency). As type I is the mildest form of OI, it demonstrates the fact that in many cases of mutations involving genes that encode structural polypeptides or proteins, it can be better to have no gene product than to have an abnormal one, a concept which is being explored in the treatment of structural protein disorders by attempting to silence the mutated gene.
In other cases, the variant leads to production of structurally abnormal pro-α1 or pro-α2 chains that are subject to aberrant post translational modification, or inability to associate properly with the other pro-chains to form a strong helix. When a single mutation on only one allele results in a structurally abnormal protein that actually interferes with the functioning of the residual normal proteins, this is termed a “ dominant negative ” effect. As a general rule, the earlier the altered mRNA codon appears in the translation process, the more abnormal is the resulting prochain structure, and the greater is the degree of compromise of collagen strength and function within connective tissues. These types of mutations, which result in the synthesis of structurally abnormal products that interfere with collagen structure and function, are the basis for clinical abnormalities found in types II to IV OI. The phenomenon of excessive posttranslational modification of a structurally abnormal gene product is also seen in some types of Ehlers-Danlos syndrome (EDS; see the Ehlers-Danlos Syndrome section, later).
See Table 1.10 for examples of some hereditary connective tissue disorders.
| Diagnosis | Inheritance | Molecular Basis |
|---|---|---|
| Stickler syndrome | AD | COL2A1 gene |
| COL11A1 gene | ||
| COL11A2 gene | ||
| COL9A1 gene | ||
| vEDS | AD | COL3A1 gene |
| kEDS- PLOD1 | AR | PLOD1 gene |
| Beals contractural arachnodactyly | AD | FBN2 gene |
| Homocysteinemia | AR | Defect in cobalamin synthesis |
| Arterial tortuosity syndrome | AR | SLC2A10 gene |
| MASS phenotype | AD | FBN1 gene |
| Loeys-Dietz syndrome | AD | TGFBR1 gene |
| TGFBR2 gene | ||
| Familial aortic aneurysm | AD | ACTA2 gene |
| MYH11 gene | ||
| Klinefelter syndrome (47,XXY) or triple X syndrome (47, XXX) | Chromosomal | Chromosomal |
| Fragile X syndrome | X-linked | FMR1 gene |
| Shprintzen-Goldberg syndrome | AD | SKI gene |
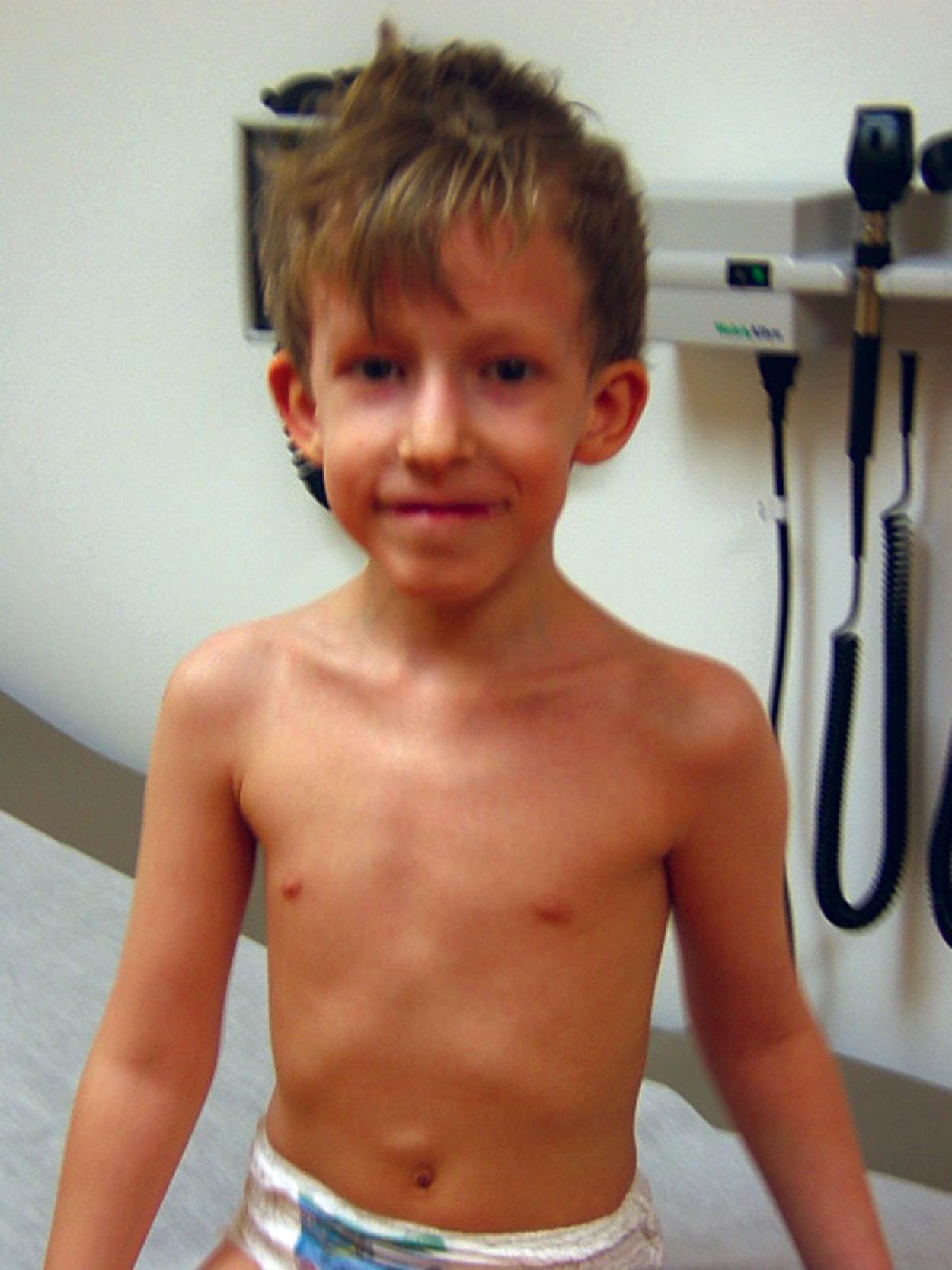
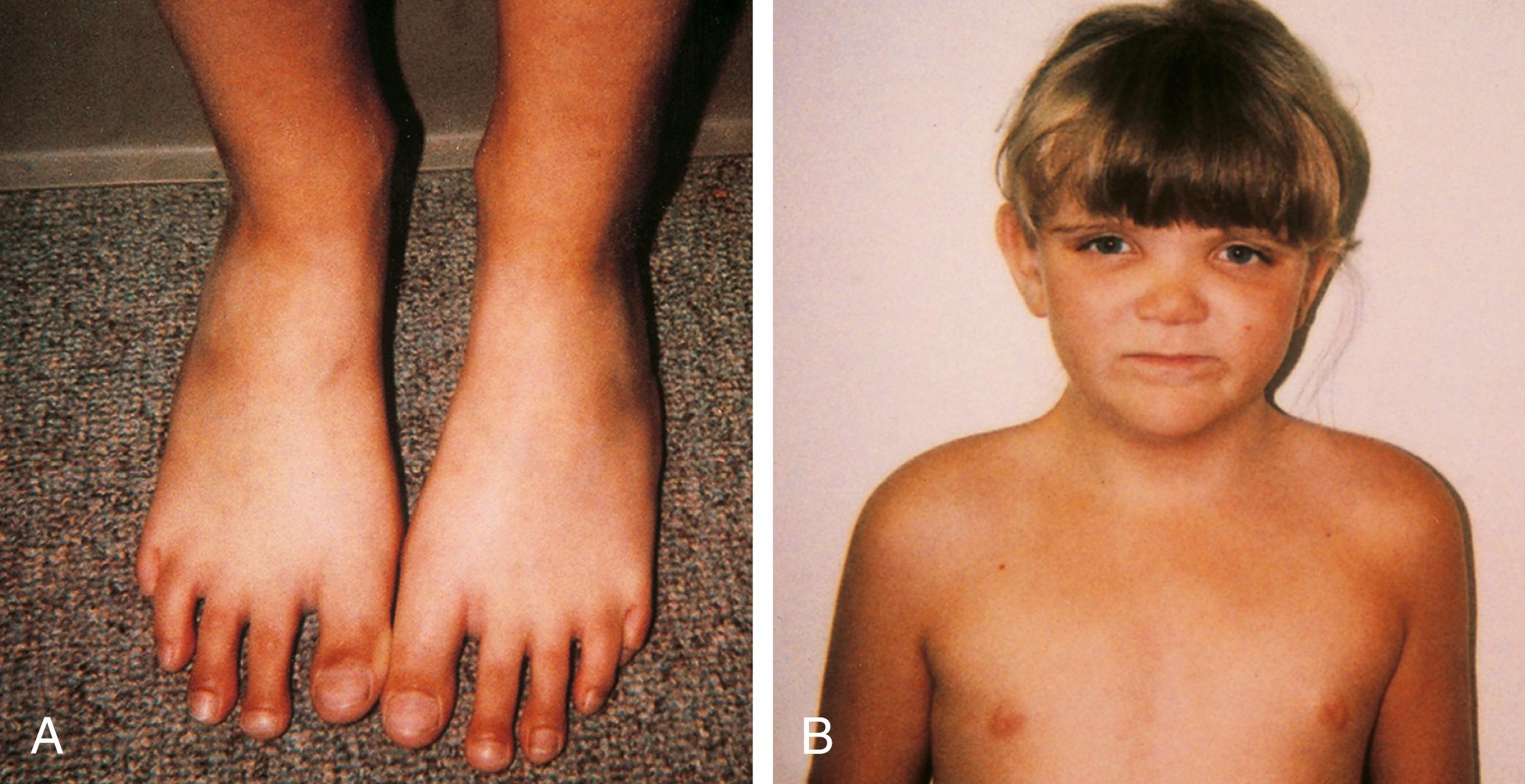
Marfan syndrome is a genetic disorder of connective tissue that is inherited as an autosomal dominant trait, although approximately 25% to 30% of cases represent new mutations. The site of the genetic abnormality or mutation is the fibrillin gene (FBN1) located at band 15q21.1 on chromosome 15. As a result, the molecular structure of the protein fibrillin, an intrinsic component of connective tissue, is abnormal. Clinical consequences are most notable in the musculoskeletal, cardiovascular, and ocular systems and in the dura. Approximately 70% of cases are familial. Classic phenotypic findings include arachnodactyly (see Fig. 1.32A and B ); elbow contractures with other joint hyperextensibility due to ligamentous laxity (see Chapter 5 ); tall stature with long, thin extremities (termed a “Marfanoid” habitus); a decreased upper-to-lower segment ratio; an arm span to height ratio that exceeds 1.05; and moderate to severe pectus excavatum or carinatum (see Fig. 1.32C ). Pes planus and thoracolumbar kyphoscoliosis are other common skeletal features (see Fig. 1.32D ). A defect in the suspensory ligaments of the eye is responsible for subluxation of the lens (seen in 50% to 60% by 10 years old), which is usually displaced in an upward direction. Myopia and astigmatism are common, and affected individuals are also at risk for developing glaucoma, cataracts, and retinal detachment in adulthood. Mitral valve prolapse may progress to mitral insufficiency (at times associated with arrhythmias). Of great concern is progressive aneurysmal dilatation of the ascending aorta and, less commonly, the thoracic or abdominal aorta. The latter is the major source of morbidity and mortality because it can result in acute dissection and death. Dural ectasia in the lumbosacral region, assessed by computed tomography (CT) or magnetic resonance imaging (MRI) of the spine, is observed in 65% of cases. The presence of a high arched palate is common. The incidence of hernias, both inguinal and femoral, is increased, and patients often have striae of the skin in unusual places such as the shoulder. Although most Marfan individuals are of normal intelligence, an occasional patient may have learning disabilities, which should prompt additional investigation.
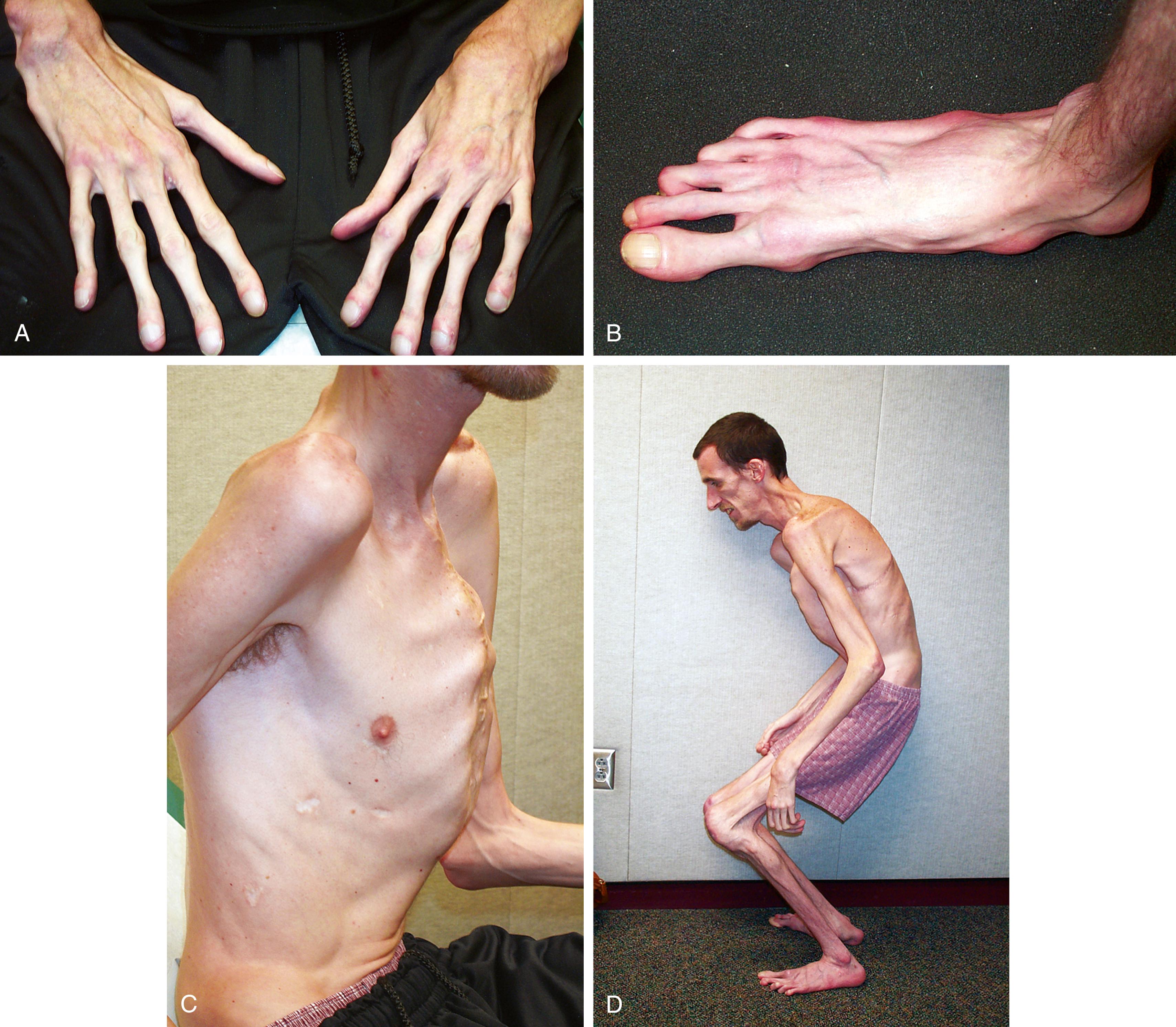
The disorder is currently primarily suspected on clinical grounds, but family history and multiorgan manifestations are variable and can have age-dependent expressivity. Attempts have been made to create diagnostic criteria by establishing major and minor manifestations (first established in Berlin; Beighton et al., 1988) and revised as the Ghent criteria, most recently revised in 2010 ( ). The diagnostic criteria are based on cardiovascular, ocular, and skeletal features; the presence of a dural ectasia; and family history and have approximately 92% sensitivity in adults. These revisions have placed an increasing emphasis on the cardinal features of Marfan syndrome. Because it takes time for a number of the major abnormalities to develop or to become clinically evident, a firm diagnosis is generally impossible in early childhood without molecular testing, especially in the absence of a positive family history. Molecular testing is being used more frequently in children with an emerging clinical phenotype, especially in the absence of family history and to assess for genotype-phenotype correlations. A molecular diagnosis also provides for the diagnosis of family members who might have variable expressivity and fail to meet the diagnostic criteria on their own. The recurrence risk for affected individuals to their offspring is 50%; prenatal diagnosis is possible if the underlying mutation is known. Pregnant women with Marfan Syndrome require high risk management for cardiovascular complications and potential teratogenicity of some medication treatments.
The differential diagnosis of Marfan syndrome includes Loeys-Dietz syndrome (LDS); Beals congenital contractural arachnodactyly (see Figs. 1.33 through 1.35 ); homocystinuria; the MASS phenotype; familial ectopia lentis; Klinefelter syndrome (47,XXY), triple X syndrome (47,XXX), and many syndromes characterized by joint hypermobility, such as Stickler syndrome (see Figs. 1.38 and 1.39 ), classic EDS (cEDS) (see Fig. 1.36 ), and vascular EDS (vEDS); familial thoracic aortic aneurysm and aortic dissection (TAAD); neuromuscular disorders; fragile X syndrome; and some of the rare dysmorphologic entities, such as Shprintzen-Goldberg syndrome.

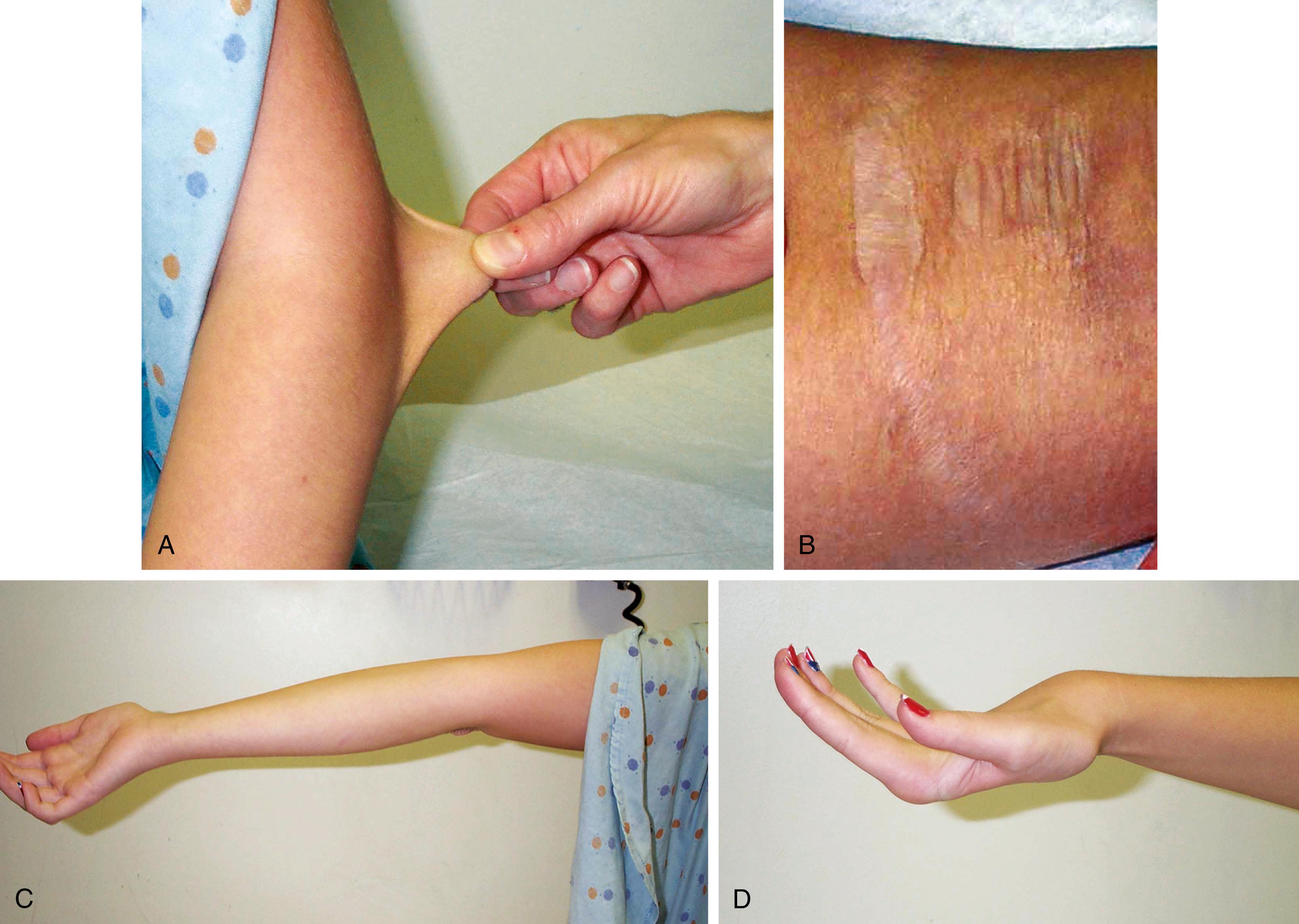
When the diagnosis of Marfan syndrome is strongly suspected or confirmed, patients should be monitored closely during growth spurts for signs of onset and progression of kyphoscoliosis; in addition, they should undergo regular ophthalmologic evaluations, and have regular echocardiograms and electrocardiograms. When aortic dilatation is detected, administration of β-blockers can slow progression by decreasing blood pressure and the force of myocardial contractions. Research has found transforming growth factor-β (TGF-β) to be involved in the formation of aortic aneurysms. Losartan, an angiotensin II type 1 receptor blocker, inhibited TGF-β in a mouse model of Marfan syndrome and has now become standard of care for pediatric and adult Marfan and LDS patients. Subacute bacterial endocarditis (SBE) prophylaxis may or may not be indicated for patients with evidence of cardiovascular involvement. Patients also should be cautioned to avoid weightlifting and contact sports.
LDS is a more aggressive connective tissue disorder than Marfan syndrome; it is characterized by craniofacial, cutaneous, and skeletal and vascular manifestations, as well as aneurysms and dissection of the aorta (in the root, thoracic, and/or abdominal regions) and cerebral vessels (see Fig. 1.33 ). Tortuosity of blood vessels and heart defects, involving the bicuspid aortic valves and atrial septal defects, may be observed. Bifid uvula is a common finding. Translucent skin and organ rupture, specifically in postpartum females, have been reported. Pathogenic variants in TGF-β receptor type 1, TGFBR1, and TGFBR2 result in LDS. Treatment strategies and recommendations are very similar to Marfan syndrome described above.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here