Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
This work was partially supported by the Spanish National Bioinformatics Institute (INB) and grants BIO2012-40205, SAF2011-29530 (MINECO, Spain) and COST Action #BM1204: EU_Pancreas (European Cooperation in Science and Technology).
The technology revolution by the Human Genome Project from 1986 to 2003 has propelled the field of bioinformatics into a new era, with the formidable challenge of organizing, classifying, making available, and interpreting complex genomic data within the context of large biological databases and repositories containing the accumulated information.
The major advances in experimental methods for genome characterization based on deoxyribonucleic acid (DNA)/ribonucleic acid (RNA) microarrays and DNA sequencing—for example, capillary-based DNA Sanger sequencing and, more recently, next-generation sequencing (NGS)—make analysis mutations, gene expression, and copy number alterations possible in a large number of cancer genomes . Indeed, 21st-century sequencing-based experiments generate substantially more data and are more broadly applicable than microarray technology, allowing for various novel functional assays, including quantification of protein–DNA binding or histone modifications (chromatin immunoprecipitation followed by sequencing, ChIP-seq), transcript levels using RNA sequencing (RNA-seq), and genome (WGS) and exome sequencing (WES) variant discovery.
New genomic technology has come at a cost, however, resulting in a greater challenge for associated bioinformatics analyses. The fast development of bioinformatics and the complex combination of related biology, computer science, and information technology often make it difficult for biomedical researchers to use the available technology to its fullest and, in many cases, even to select the appropriate tools and computational resources.
This chapter provides an overview of the high-throughput genomics technologies (with an emphasis in NGS), the data currently available (cancer-related databases) and the types of bioinformatic analyses that need to be applied. It emphasizes the specific challenges posed by the analysis of pancreatic samples and provides specific examples.
Standardized protocols for sample quality are essential to ensure reproducible results and comparability. In general, clinical sample experiments are complicated due to the differences in sample quantity, quality and purity. Tumor samples often include substantial fractions of necrotic or apoptotic cells as well as a mixture of malignant and nonmalignant cells. Also, nucleic acids isolated from cancer are often of lower quality than those purified from peripheral blood. Along this line, tumors may be highly heterogeneous and composed of different clones with different genomes . Furthermore, the control samples are also problematic because peripheral blood provides only an imperfect reference and surgical resections are difficult to obtain. Bioinformatics methods are being designed to alleviate most of these issues, but they still impose constraints that have to be taken into account during the organization and interpretation of cancer genome projects.
Aside from the pancreatic tissue samples, cellular components and their function pose additional challenges to the analysis. The pancreas is composed of three major cell types (acinar, ductal, and endocrine). The acinar component is the largest in normal tissue, accounting for about 80% of all cells. Therefore, a comparison of different origin tumors with the normal pancreas is not fully suited per se. Moreover, pancreatic tumors are characterized by a massive desmoplastic reaction and “contamination” from inflammatory/stromal components, which often result in a tumor mass that contains around 38% (ranging from 5% to 85%) of cancer cells . Unlike other neoplasms, histological evaluation of the cellular composition of the specimens used for analyte isolation is essential. In addition, the exocrine pancreas produces large amounts of hydrolytic enzymes, which prevent quality samples from being obtained for analysis. Appropriate controls should be used to examine analyte degradation. It is a fact that an RNA integrity number (RIN) score higher than 7, as recommended for RNA-seq experiments, is hard to obtain in human pancreas samples.
The purity and quality of the material (DNA/RNA) required will have a decisive influence on the quality of the raw data obtained and should be taken into account during the design of the analysis, selection of algorithms, and the follow-up interpretation of the results. The basic bioinformatics approaches applied to the initial raw data are described in detail in the following section.
The development of the microarray technology at the end of the past century was a revolution in the molecular biology field. Microarrays allowed multiple hypotheses to be interrogated simultaneously with robust methods, thereby leading to an application in gene discovery, gene regulation, biomarker determination, and disease classification. Microarrays have been used widely in research, and they commonly are found in the facilities of many academic institutions and biotechnology companies.
Microarrays are hybridization based and commonly are used to measure the binding of a nucleic acid analyte on the basis of sequence complementarity. This allows both analysis of expression and genotyping. cDNA (two colors) and oligonucleotide (single color) microarrays are the two main microarrays platforms and both have been used widely. cDNA microarrays are useful to measure transcript abundance and are based in printed cDNA with size ranging from a few 100 bases to several kilobases. In two-color arrays, the test and reference samples are labeled with fluorescent Cy5 or Cy3 dyes, using reverse transcriptase, and subsequently are hybridized. The slides are scanned to measure fluorescence, and the signal is relative to the abundance hybridized transcripts. In oligonucleotide microarrays, the probes are directed synthesized on glass slides using photolithography technology. The probes are usually 9–50 nucleotide oligonucleotides that hybridize with samples labeled with biotin or Cy3 dye (single-color arrays).
The first step in the analysis of microarray data is normalization, which is aimed at compensating differences in labeling, hybridization, and detection methods. Data normalization is essential for comparison of different experiments. The selection of the appropriate method depends on the type of array and expected biases. Total intensity normalization assumes that the total hybridization intensities summed over all elements in the arrays should be the same for each sample. In addition, there are a number of alternative approaches to the total intensity normalization method, including linear regression analysis, log centering, rank invariant methods, and others. Because these methods can have a systematic dependence on intensity, the effect of which is often nonlinear and can vary from different slides, locally weighted linear regression normalization has been proposed as a method to remove intensity-dependent effects, taking into account individual slides to remove slide-dependent dye effect .
Most normalization algorithms can be applied to the entire data set (global normalization) or to some subset of the data (local normalization). Local normalization helps to correct spatial variations in the array, such as variability in slide surface or slight differences in hybridization conditions across the arrays. Then the variability between regions of an array or between arrays should be corrected so that their variance is the same, normally achieved by adjusting the log2 (ratio) measurements .
Housekeeping genes frequently have been used to normalize microarray expression data under the assumption that they display stable levels across samples. This is not always the case, however, leading to erroneous conclusions as shown by Welsh et al. and Yu et al. . Other strategies have been proposed to overcome the housekeeping limitations based on identifying genes that are not expressed differentially across different biological samples in the same data set, to normalize the data. A major normalization effort should be made using standardized spike-in controls of known concentration, defined length, and guanine–cytosine (GC) content . Finally, visual inspection is recommended using box plots, scatter plots, or MA plots—plots of the distribution of the Cy5/Cy3 intensity ratio (‘M’) versus the average intensity (‘A’)—to identify possible errors introduced during the normalization procedure. Even though many normalization algorithms have been developed, the Limma package has gained wide acceptance and includes all of the necessary tools for the analysis of the different types of array-based experiments. Limma is part of Bioconductor ( http://www.bioconductor.org/ ), an open-source package based on the R programming language for the analysis of high-throughput genomic data, including microarrays.
After data normalization, the analysis of expression microarrays can be carried out with supervised or unsupervised methods. Supervised methods identify differential gene expression (DGE) patterns between samples of known phenotypes—for example, cells that are exposed or not exposed to experimental manipulation. A number of statistical tests are applied—such as the t -test, Wilcoxon rank-sum test, or significant analysis of microarray—to identify the DGE between two groups or tests based on analysis of variance to identify differential expression between multiple groups. Microarrays test multiple hypotheses in a single experiment and can produce hundreds of false-positive results. These false positives that result from multiple comparisons are controlled by family-wise error rate or the false discovery rate (FDR) estimations. The first represents the probability of having at least one false-positive result for all the tests and the second is less stringent and provides the expected proportion of false positives among the significant results.
Unsupervised methods group samples or genes based on their expression distance, without using information about the associated phenotypes. The most common method used is hierarchical clustering, which groups the samples that have similar expression patterns, genes that are highly correlated, or both, producing dendrograms in which the length of the branches is inversely proportional to the similarity between samples or genes. Other unsupervised methods are K-means, principal component analysis, or self-organizing maps.
Moreover, the unit of analysis can be “gene modules” instead of individual genes, as the latter can be grouped based on previous biological knowledge. The genes can be grouped according to biological pathway, motif sharing, or tissue expression. This kind of analysis has been termed “functional analysis”. Three types of methods follow this strategy. One is the singular enrichment analysis (SEA), which is the best strategy established for enrichment analysis and is based on a preselected list of genes defined by the user. This is measured by different statistics, such as chi-squared, Fisher’s, binomial, or hypergeometric tests. Another is the Gene-Set Enrichment Analysis (GSEA), which is based on a ranked list of genes and Kolmogorov–Smirnov statistics. This method has the advantage of not requiring arbitrary cutoffs. The third method is the modular enrichment analysis, which incorporates extra network discovery algorithms into the SEA methodology.
In the case of microarrays used for genotyping, the software provided by the manufacturers normally is used for the normalization and genotype analysis step. Currently, microarrays are able to genotype more than a million single-nucleotide polymorphisms (SNPs) simultaneously. The first step is to summarize the probe intensities for each SNP, followed by a call based on the summarized intensities. There are three possible genotypes (assuming diploidy): AA, BB (homozygous), and AB (heterozygous), where A and B denote the two possible alleles. Further steps include linkage disequilibrium and phasing, where alleles at two or more loci appear together in the same individual more often than would be expected by chance.
Genome-wide association studies (GWAS) use germline DNA to identify genetic variants that are more common in individuals with a given phenotype than in the control population. They provide a powerful tool to analyze genetic variation but are limited by the false-positive rate derived from the large number of comparisons performed. To acquire statistical significance—in the case of common, low-penetrance, alleles—large numbers of affected (cases) and unaffected (controls) individuals are needed. In microarray genotyping, the signal intensity is related with the DNA amount harboring the region interrogated by the probe. Therefore, the probe intensity can be used for further analyses, including DNA copy changes, the detection of loss of heterozygosity (LOH) and uniparental disomies, and other structural alterations. Algorithms for copy number—such as WaviCGH —follow similar steps, including the summarization of the intensity of consecutive probes (2-40) into a single measure, followed by segmentation to infer chromosomal segments of constant copy number, the calling of gains and losses regions, and the identification of minimal common regions over a set of samples.
Microarrays have been used widely in the cancer field for more than a decade for detection of biomarkers, sample classification, response to treatment, and drug screening. Unlike with other tumor types, the number of data sets and samples available in pancreatic cancer is limited. For example, across the 715 microarray data sets and 87,633 samples included in Oncomine (see description in databases and resources section) only 29 data sets with 606 samples referred to pancreatic cancer. In contrast, other frequent tumors (such as colorectal, breast, lung, or brain) are represented by more than twice the number of the data sets, with up to 132 data sets, which include 14,277 samples, in the breast. This reflects the difficulty of accessing pancreatic samples and obtaining sufficient quantity and quality compared with other tumor types as noted in the previous sections.
The earliest studies using expression microarrays focused on the use of gene expression profiles to characterize pancreatic ductal adenocarcinoma (PDAC) . These studies identified sets of genes differentially expressed between PDAC and normal samples ranging from 75 to 587 genes. Grutzmann et al. carried out the first meta-analysis, which showed 568 deregulated genes in pancreatic cancer, of which only 22% had been described previously. Following these studies, Badea et al. defined their own DGE set in 36 pancreatic tumor tissues, and compared the list with previous results on pancreatic cancer and microarrays from 25 different publications, to define a list of target genes involved in pancreatic cancer. This strategy allowed the identification of 135 genes of the 239 from its data set in any of the other studies, some of them with prognosis and survival implications. Collisson et al. (128) were the first to use transcriptomic data to identify subtypes of pancreatic adenocarcinoma, characterized on the basis of their gene expression profiles, with potential implications for therapeutic response . These studies need to be validated in independent series. Expression microarrays also have been applied to assess the drug response of pancreatic cancer cell lines, primary cultures, or xenografts. These approaches have identified genes such as Rrm1, Top2a, Casp3, and others that have shown resistance to gemcitabine, the standard treatment for advanced pancreatic cancer. A review on gene expression profiling and pancreatic cancer can be consulted in reference . Recently, Gadaleta et al. performed the most significant integrated analysis in pancreas cancer to date using microarrays expression data, for a total of 309 samples from different studies and sources (cancer pancreas samples, cell lines, xenografts) using the same microarray platform. The main findings of this study pointed out that normal samples adjacent to tumors often display transcriptomic changes, and the xenografts and cell line models do not fully recapitulate the transcriptome of primary tumors (detailed results can be consulted in the Pancreatic Expression Database (PED)). This may explain the differences between studies and the difficulty in moving gene expression profiles to the clinic. Another interesting resource for pancreatic cancer studies is the microarray transcriptome characterization in islets of healthy human donors carried out by Dorrel et al. . These authors used cell type-specific surface-reactive antibodies to capture dispersed single cells and to characterize the transcriptome of alpha, beta, large-duct, small-duct, and acinar cells.
The largest GWAS conducted in pancreatic cancer was reported by Petersen et al. . This report included 3851 cases and 3934 controls from 20 studies and identified eight SNPs overlapping with three regions associated with pancreatic cancer risk (1q32.1, 5p15.33, and 13q22.1). The region 1q32.1 includes five specific SNPs associated with pancreatic cancer susceptibility for gene LRH1/NR5A2 , an “orphan” nuclear receptor critical in development. Another SNP identified in 5p15.33 was placed in CPTM1L-TERT locus, genes that have been implied in carcinogenesis. The region 13q22.1 is a large nongenic region with two associated SNPs that appear to be specific to pancreatic cancer. As commented previously, genotyping microarrays have been used widely in pancreatic cancer to assess copy number aberrations. Several amplifications have been described in different studies, but those related to oncogenes, such as KRAS, MYC, or AKT2 , have been described in multiple cases, as well as deletions affecting tumor suppressors, such as TP53, CDKN2A , and SMAD4 . Unfortunately, until now neither RNA- nor DNA-based studies have provided the basis for improved diagnostic or predictive tools. Lack of replication studies together with the challenges related to the disease and the need to obtain samples using invasive procedures contribute to this slow progress. In fact, access to clinical samples is difficult because only 20% of cases with pancreatic cancer undergo surgery and most patients are very sick at the time of diagnosis and have an extremely short life expectancy.
Next-generation sequencing (NGS) is used for the identification of protein binding to chromatin, quantification of RNA levels, and identification of mutations, as well as for other applications. Some bioinformatic processing of the data is common to all of these, as seen in the schematic workflow provided in Figure 5.1 .
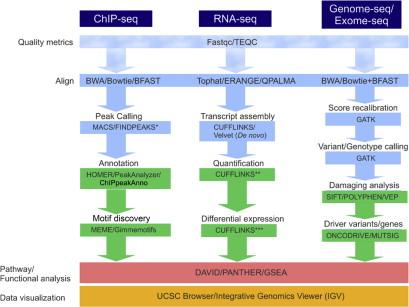
One of the earliest applications of NGS is ChIP-seq. This technique generates genome-wide profiling of DNA-bound proteins by sequencing the DNA fragments hybridized to the proteins recognized by the antibodies. Proteins are therefore in contact with the DNA directly or as part of larger protein complexes . ChIP-seq outperforms previous techniques (e.g., ChIP–ChIP microarrays) in terms of resolution, coverage, and dynamic range. It also presents fewer artifacts, for small- and large-scale approaches, including the first genome-scale view of DNA–protein interactions , and for these reasons, it has become widely used.
Briefly, the DNA-binding protein is cross-linked to DNA sheared into small fragments (200–600 bp) and immunoprecipitated with an antibody specific to the protein of interest. The immunoprecipitated DNA fragments then are used as the input for the sequencing library preparation protocol and finally are sequenced. Although the Illumina/Solexa Genome analyzers dominate the NGS market, multiple platforms have been developed.
Guidelines for good practices and quality metrics for ChIP-seq experiments have been developed by the Encyclopedia of DNA Elements (ENCODE) consortium . In brief, ideally the objective is (1) to obtain ≥10 million uniquely mapping reads per replicate experiment, (2) to generate and sequence a control ChIP library for each experiment (cell type, tissue, or embryo collection), and (3) to perform experiments at least twice to ensure reproducibility.
Become a Clinical Tree membership for Full access and enjoy Unlimited articles
If you are a member. Log in here